You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002602_01765
You are here: Home > Sequence: MGYG000002602_01765
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
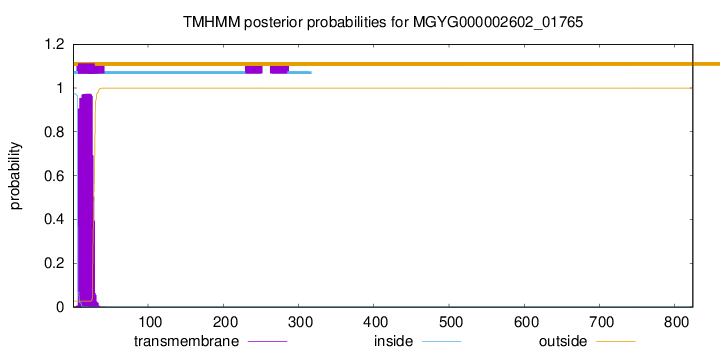
TMHMM annotations
Basic Information help
| Species | CAG-274 sp900545305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; CAG-274; CAG-274 sp900545305 | |||||||||||
| CAZyme ID | MGYG000002602_01765 | |||||||||||
| CAZy Family | CBM77 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 294; End: 2768 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 420 | 797 | 1.4e-54 | 0.8186666666666667 |
| CBM77 | 56 | 144 | 8.3e-18 | 0.8155339805825242 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18283 | CBM77 | 1.51e-16 | 56 | 151 | 17 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBI14069.1 | 3.88e-179 | 212 | 824 | 33 | 606 |
| CBZ48778.1 | 1.23e-177 | 212 | 824 | 33 | 606 |
| BAK28435.1 | 1.23e-177 | 212 | 824 | 33 | 606 |
| QKI00169.1 | 1.23e-177 | 212 | 824 | 33 | 606 |
| QWX86240.1 | 1.23e-177 | 212 | 824 | 33 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 2.73e-14 | 498 | 756 | 109 | 298 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 2.37e-14 | 405 | 756 | 47 | 323 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.74e-13 | 498 | 756 | 134 | 323 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000494 | 0.185436 | 0.813735 | 0.000123 | 0.000119 | 0.000083 |