You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002603_00198
You are here: Home > Sequence: MGYG000002603_00198
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900767615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900767615 | |||||||||||
| CAZyme ID | MGYG000002603_00198 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 248522; End: 250006 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 2.40e-11 | 198 | 369 | 9 | 178 | pectinesterase |
| PLN02176 | PLN02176 | 9.92e-10 | 196 | 401 | 41 | 248 | putative pectinesterase |
| PLN02682 | PLN02682 | 1.15e-09 | 182 | 396 | 58 | 270 | pectinesterase family protein |
| PLN02995 | PLN02995 | 2.57e-08 | 175 | 371 | 204 | 390 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02488 | PLN02488 | 5.79e-08 | 193 | 486 | 196 | 487 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65269.1 | 0.0 | 1 | 494 | 38 | 531 |
| QVJ81037.1 | 6.48e-226 | 2 | 492 | 4 | 497 |
| ADE82670.1 | 6.60e-225 | 2 | 492 | 20 | 513 |
| QJD94164.1 | 1.32e-164 | 27 | 492 | 25 | 486 |
| QBI04917.1 | 6.41e-157 | 23 | 492 | 9 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C1C_A | 4.90e-06 | 184 | 491 | 2 | 295 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 4.90e-06 | 184 | 491 | 2 | 295 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 1GQ8_A | 9.47e-06 | 196 | 371 | 9 | 172 | Pectinmethylesterase from Carrot [Daucus carota] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LVQ0 | 5.10e-10 | 191 | 398 | 2 | 211 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| P24791 | 1.85e-09 | 179 | 428 | 63 | 353 | Pectinesterase OS=Ralstonia solanacearum OX=305 GN=pme PE=3 SV=1 |
| Q9FM79 | 2.31e-09 | 137 | 431 | 15 | 316 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9FHN5 | 2.57e-09 | 175 | 489 | 201 | 511 | Probable pectinesterase/pectinesterase inhibitor 59 OS=Arabidopsis thaliana OX=3702 GN=PME59 PE=2 SV=1 |
| Q3EAY9 | 1.72e-08 | 179 | 489 | 170 | 478 | Probable pectinesterase 30 OS=Arabidopsis thaliana OX=3702 GN=PME30 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
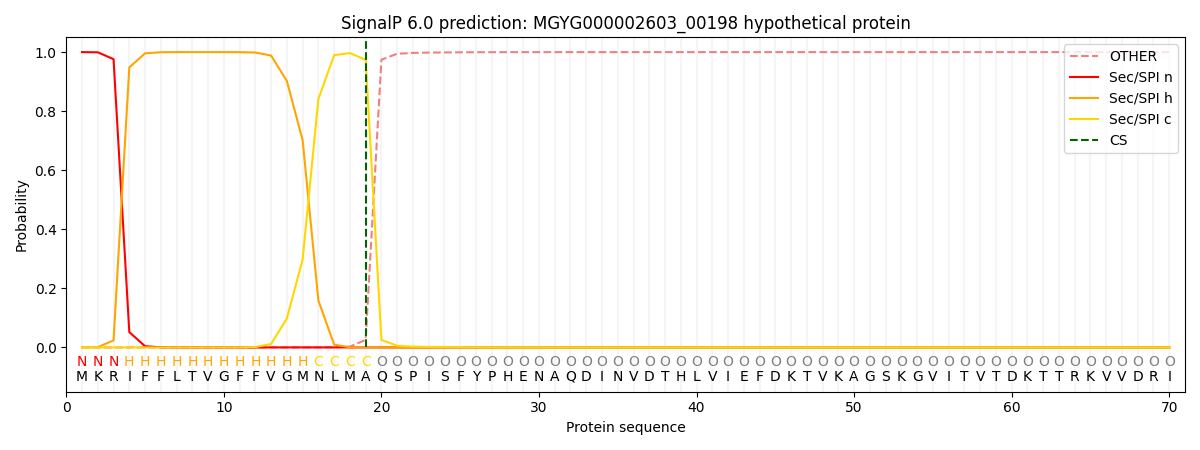
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000407 | 0.998834 | 0.000237 | 0.000172 | 0.000161 | 0.000151 |
