You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002603_01325
You are here: Home > Sequence: MGYG000002603_01325
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900767615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900767615 | |||||||||||
| CAZyme ID | MGYG000002603_01325 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41068; End: 43425 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 475 | 757 | 1.5e-100 | 0.9818840579710145 |
| GH26 | 99 | 428 | 7.8e-88 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.55e-50 | 478 | 756 | 19 | 266 | Cellulase (glycosyl hydrolase family 5). |
| pfam02156 | Glyco_hydro_26 | 2.61e-43 | 99 | 428 | 2 | 311 | Glycosyl hydrolase family 26. |
| COG2730 | BglC | 4.83e-20 | 444 | 717 | 48 | 323 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG4124 | ManB2 | 5.93e-14 | 219 | 350 | 145 | 260 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65320.1 | 0.0 | 1 | 785 | 1 | 786 |
| BCS86234.1 | 0.0 | 1 | 785 | 1 | 776 |
| AGH13958.1 | 0.0 | 1 | 784 | 1 | 775 |
| AHG56239.1 | 0.0 | 2 | 784 | 150 | 923 |
| AAC97596.1 | 0.0 | 2 | 784 | 150 | 923 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D2W_A | 0.0 | 1 | 784 | 1 | 775 | Crystalstructure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14],6D2W_B Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14] |
| 6D2X_A | 6.52e-172 | 95 | 432 | 2 | 337 | Crystalstructure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii [Prevotella bryantii B14] |
| 5D9N_A | 1.31e-135 | 429 | 784 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
| 5D9M_A | 1.04e-134 | 429 | 784 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9M_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9O_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii],5D9O_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii] |
| 3VDH_A | 3.53e-129 | 429 | 784 | 2 | 352 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii],3VDH_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10477 | 8.02e-52 | 439 | 781 | 56 | 383 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P17901 | 1.89e-51 | 434 | 781 | 41 | 398 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P16216 | 8.78e-50 | 428 | 780 | 57 | 395 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
| Q12647 | 2.54e-48 | 439 | 781 | 24 | 361 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P23661 | 1.08e-47 | 439 | 781 | 70 | 398 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
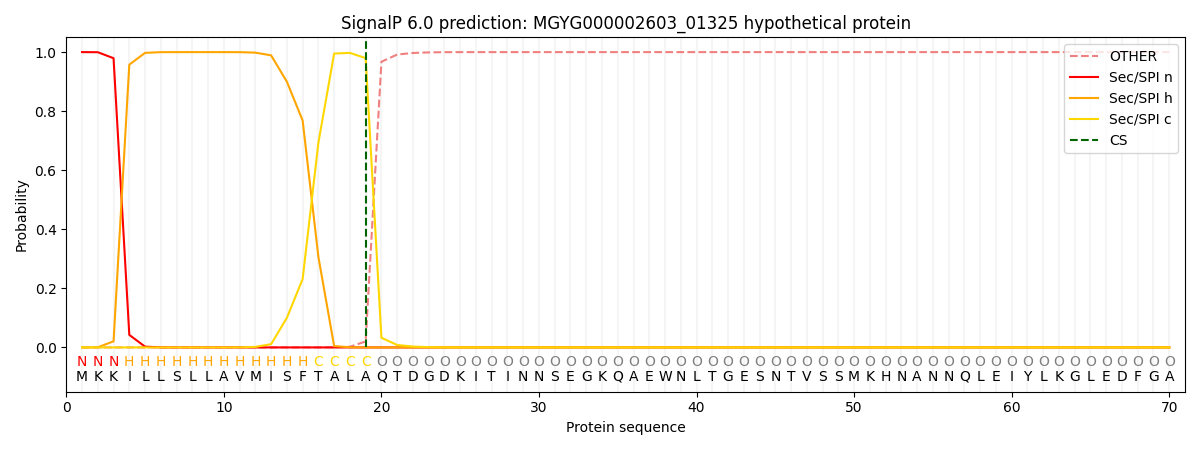
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000224 | 0.999102 | 0.000183 | 0.000166 | 0.000157 | 0.000148 |
