You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002603_01366
You are here: Home > Sequence: MGYG000002603_01366
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900767615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900767615 | |||||||||||
| CAZyme ID | MGYG000002603_01366 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 103279; End: 106239 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 29 | 712 | 1.8e-77 | 0.6808510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.92e-22 | 122 | 472 | 63 | 427 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.72e-18 | 120 | 452 | 63 | 421 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.21e-11 | 126 | 448 | 113 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 2.67e-11 | 120 | 211 | 63 | 146 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 8.59e-09 | 231 | 321 | 2 | 105 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADE82591.1 | 0.0 | 15 | 985 | 5 | 900 |
| QVJ80649.1 | 0.0 | 15 | 985 | 5 | 900 |
| AGB28395.1 | 0.0 | 25 | 985 | 24 | 976 |
| BCS84402.1 | 0.0 | 32 | 985 | 11 | 899 |
| QIU94545.1 | 0.0 | 26 | 985 | 19 | 957 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XXW_A | 2.59e-18 | 118 | 450 | 68 | 418 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6U7I_A | 8.84e-16 | 118 | 449 | 61 | 413 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 4JKK_A | 1.71e-13 | 74 | 452 | 12 | 415 | CrystalStructure of Streptococcus agalactiae beta-glucuronidase in space group I222 [Streptococcus agalactiae 2603V/R],4JKL_A Crystal Structure of Streptococcus agalactiae beta-glucuronidase in space group P21212 [Streptococcus agalactiae 2603V/R],4JKL_B Crystal Structure of Streptococcus agalactiae beta-glucuronidase in space group P21212 [Streptococcus agalactiae 2603V/R] |
| 3K4A_A | 2.27e-13 | 104 | 449 | 49 | 416 | Crystalstructure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12],3K4A_B Crystal structure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12] |
| 6LEM_B | 3.92e-13 | 104 | 449 | 45 | 412 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O33815 | 2.25e-14 | 154 | 411 | 142 | 387 | Beta-galactosidase OS=Staphylococcus xylosus OX=1288 GN=lacZ PE=3 SV=1 |
| P26257 | 2.79e-13 | 79 | 442 | 8 | 382 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| P05804 | 2.15e-12 | 104 | 449 | 47 | 414 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| Q48846 | 3.87e-12 | 137 | 467 | 144 | 485 | Beta-galactosidase large subunit OS=Latilactobacillus sakei OX=1599 GN=lacL PE=3 SV=1 |
| Q04F24 | 6.17e-11 | 154 | 455 | 148 | 472 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
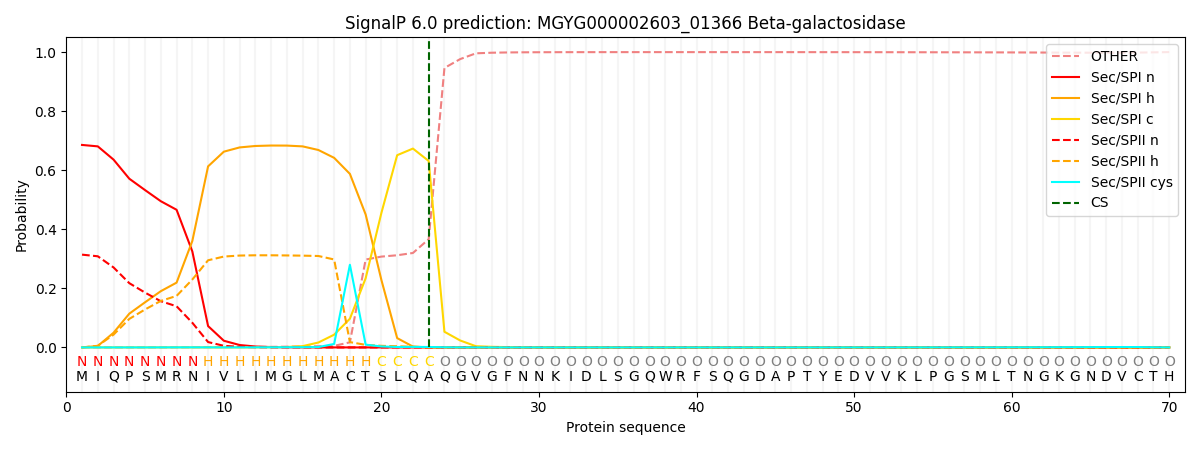
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000821 | 0.676066 | 0.322236 | 0.000339 | 0.000271 | 0.000241 |
