You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002613_00113
You are here: Home > Sequence: MGYG000002613_00113
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000002613_00113 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29707; End: 32403 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 397 | 544 | 3.7e-16 | 0.7127659574468085 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 7.46e-26 | 689 | 839 | 3 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 2.73e-15 | 429 | 522 | 1 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| cd00110 | LamG | 2.82e-08 | 685 | 829 | 1 | 150 | Laminin G domain; Laminin G-like domains are usually Ca++ mediated receptors that can have binding sites for steroids, beta1 integrins, heparin, sulfatides, fibulin-1, and alpha-dystroglycans. Proteins that contain LamG domains serve a variety of purposes including signal transduction via cell-surface steroid receptors, adhesion, migration and differentiation through mediation of cell adhesion molecules. |
| cd00161 | RICIN | 6.59e-04 | 399 | 522 | 1 | 111 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| smart00159 | PTX | 0.001 | 744 | 855 | 72 | 186 | Pentraxin / C-reactive protein / pentaxin family. This family form a doscoid pentameric structure. Human serum amyloid P demonstrates calcium-mediated ligand-binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB29926.1 | 0.0 | 4 | 882 | 5 | 892 |
| QIM09902.1 | 6.11e-216 | 54 | 847 | 29 | 829 |
| ASB47762.1 | 9.63e-216 | 43 | 850 | 51 | 845 |
| QUT88791.1 | 1.89e-214 | 6 | 842 | 19 | 844 |
| ALJ60208.1 | 3.33e-212 | 6 | 842 | 19 | 844 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A3R0A696 | 3.32e-07 | 619 | 830 | 25 | 259 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| P36905 | 1.77e-06 | 548 | 856 | 927 | 1209 | Amylopullulanase OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=apu PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
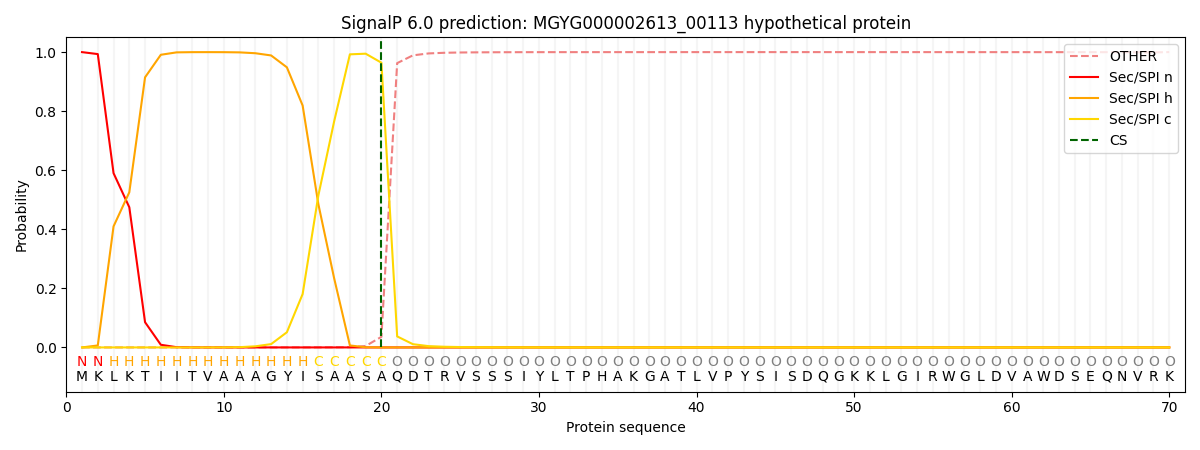
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000897 | 0.997601 | 0.000471 | 0.000375 | 0.000326 | 0.000298 |
