You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002614_00435
You are here: Home > Sequence: MGYG000002614_00435
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; | |||||||||||
| CAZyme ID | MGYG000002614_00435 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17556; End: 20543 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 38 | 865 | 7.8e-128 | 0.9574468085106383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10340 | ebgA | 1.34e-80 | 45 | 854 | 43 | 868 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 6.01e-70 | 32 | 436 | 1 | 428 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK09525 | lacZ | 1.38e-54 | 45 | 436 | 54 | 484 | beta-galactosidase. |
| PRK10150 | PRK10150 | 1.88e-47 | 46 | 414 | 15 | 417 | beta-D-glucuronidase; Provisional |
| pfam02836 | Glyco_hydro_2_C | 6.04e-37 | 283 | 596 | 1 | 300 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24892.1 | 0.0 | 21 | 995 | 16 | 982 |
| QUT50213.1 | 0.0 | 4 | 994 | 3 | 993 |
| QKH86406.1 | 0.0 | 31 | 995 | 37 | 1000 |
| QCQ48019.1 | 0.0 | 31 | 995 | 37 | 1000 |
| QCQ30210.1 | 0.0 | 31 | 995 | 37 | 1000 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 9.77e-68 | 41 | 743 | 36 | 743 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 9.84e-68 | 41 | 743 | 37 | 744 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 7.47e-60 | 46 | 637 | 51 | 655 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 7.92e-56 | 46 | 873 | 47 | 884 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 6ZJV_A | 4.91e-41 | 46 | 599 | 60 | 610 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 5.39e-67 | 41 | 743 | 37 | 744 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 3.85e-55 | 46 | 875 | 61 | 925 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A5F5U6 | 6.47e-52 | 46 | 874 | 50 | 913 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| B4S2K9 | 9.19e-52 | 30 | 761 | 34 | 813 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
| P81650 | 6.53e-50 | 30 | 830 | 34 | 858 | Beta-galactosidase OS=Pseudoalteromonas haloplanktis OX=228 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
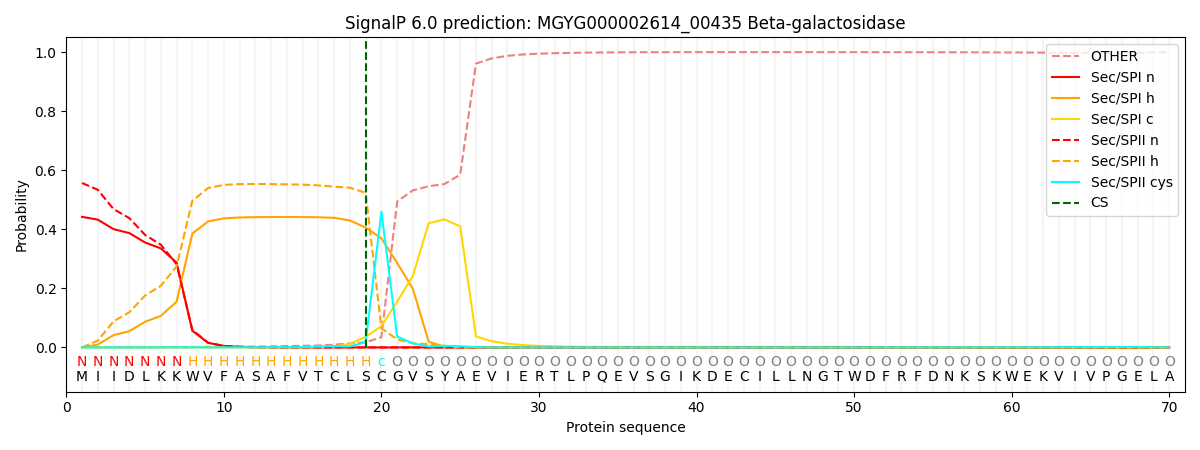
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001766 | 0.432639 | 0.564559 | 0.000534 | 0.000255 | 0.000203 |
