You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002638_01866
You are here: Home > Sequence: MGYG000002638_01866
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
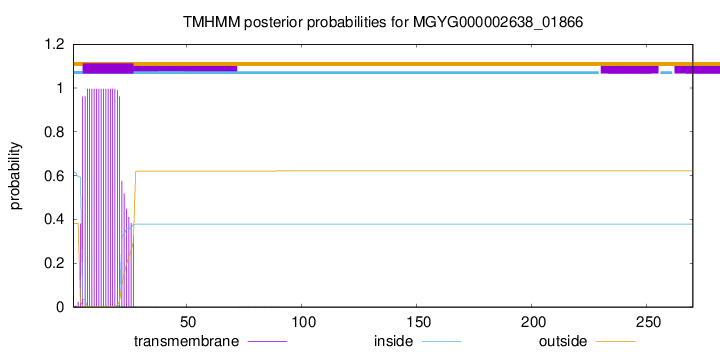
TMHMM annotations
Basic Information help
| Species | Lachnoanaerobaculum orale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoanaerobaculum; Lachnoanaerobaculum orale | |||||||||||
| CAZyme ID | MGYG000002638_01866 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9743; End: 10555 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 49 | 258 | 1.3e-59 | 0.9464285714285714 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3868 | COG3868 | 7.49e-25 | 30 | 244 | 60 | 277 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
| pfam03537 | Glyco_hydro_114 | 2.93e-23 | 49 | 144 | 18 | 112 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG2342 | COG2342 | 5.43e-20 | 3 | 244 | 1 | 254 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| cd14745 | GH66 | 5.62e-07 | 60 | 139 | 72 | 164 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| pfam14885 | GHL15 | 0.001 | 64 | 140 | 1 | 97 | Hypothetical glycosyl hydrolase family 15. GHL15 is a family of hypothetical glycoside hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYA99511.1 | 6.20e-147 | 1 | 257 | 1 | 253 |
| QUI95081.1 | 1.82e-141 | 1 | 255 | 1 | 260 |
| ASS38385.1 | 2.44e-96 | 28 | 257 | 32 | 261 |
| ACV23220.1 | 1.87e-89 | 29 | 257 | 16 | 244 |
| VEH02333.1 | 2.60e-89 | 29 | 257 | 26 | 254 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6OJ1_A | 6.24e-10 | 59 | 144 | 68 | 153 | CrystalStructure of Aspergillus fumigatus Ega3 [Aspergillus fumigatus Af293] |
| 6OJB_A | 6.96e-10 | 59 | 144 | 85 | 170 | CrystalStructure of Aspergillus fumigatus Ega3 complex with galactosamine [Aspergillus fumigatus Af293] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q58872 | 1.67e-07 | 41 | 260 | 81 | 320 | Uncharacterized protein MJ1477 OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=MJ1477 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.404724 | 0.582586 | 0.011690 | 0.000335 | 0.000273 | 0.000377 |