You are browsing environment: HUMAN GUT
MGYG000002659_00902
Basic Information
help
Species
Lineage
Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Phascolarctobacterium_A;
CAZyme ID
MGYG000002659_00902
CAZy Family
GH73
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002659
2094336
MAG
United Republic of Tanzania
Africa
Gene Location
Start: 5314;
End: 6258
Strand: +
No EC number prediction in MGYG000002659_00902.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam01832
Glucosaminidase
1.11e-08
147
213
1
77
Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O51481
7.53e-19
143
276
68
196
Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1
more
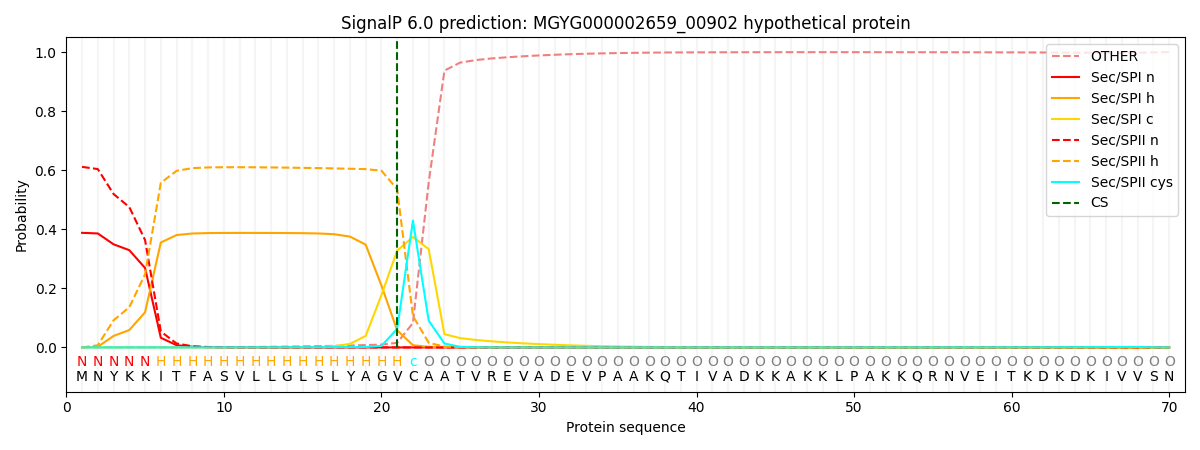
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.001504
0.379296
0.618523
0.000292
0.000192
0.000168
There is no transmembrane helices in MGYG000002659_00902.