You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002668_00312
You are here: Home > Sequence: MGYG000002668_00312
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
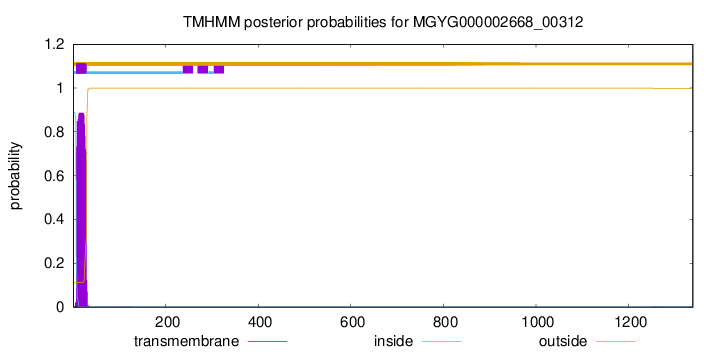
TMHMM annotations
Basic Information help
| Species | Prevotella sp900552965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900552965 | |||||||||||
| CAZyme ID | MGYG000002668_00312 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13942; End: 17961 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 81 | 258 | 1.5e-49 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15319 | PRK15319 | 5.72e-13 | 593 | 1078 | 621 | 1126 | fibronectin-binding autotransporter adhesin ShdA. |
| cd00063 | FN3 | 6.48e-11 | 438 | 507 | 4 | 77 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| PRK15319 | PRK15319 | 5.77e-10 | 542 | 985 | 894 | 1302 | fibronectin-binding autotransporter adhesin ShdA. |
| smart00060 | FN3 | 4.21e-09 | 438 | 507 | 4 | 77 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| TIGR02601 | autotrns_rpt | 2.15e-07 | 920 | 951 | 1 | 32 | autotransporter-associated beta strand repeat. This model represent a core 32-residue region of a class of bacterial protein repeat found in one to 30 copies per protein. Most proteins with a copy of this repeat have domains associated with membrane autotransporters (pfam03797, TIGR01414). The repeats occur with a periodicity of 60 to 100 residues. A pattern of sequence conservation is that every second residue is well-conserved across most of the domain. pfam05594 is based on a longer, much more poorly conserved multiple sequence alignment and hits some of the same proteins as this model with some overlap between the hit regions of the two models. It describes these repeats as likely to have a beta-helical structure. [Protein fate, Protein and peptide secretion and trafficking, Cellular processes, Pathogenesis] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV97716.1 | 8.93e-235 | 3 | 1090 | 2 | 1105 |
| QUT73817.1 | 2.08e-133 | 25 | 517 | 21 | 549 |
| QCD38445.1 | 1.92e-126 | 28 | 520 | 28 | 554 |
| QCP72135.1 | 1.49e-121 | 28 | 520 | 28 | 554 |
| QUT74519.1 | 2.56e-121 | 22 | 504 | 20 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J7K_A | 3.19e-08 | 438 | 506 | 5 | 72 | Loopgrafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_B Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_C Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_D Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_E Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_F Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_G Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_H Loop grafting onto a highly stable FN3 scaffold [synthetic construct] |
| 4U3H_A | 1.56e-07 | 438 | 506 | 17 | 84 | Crystalstructure of FN3con [synthetic construct] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0CLG7 | 7.32e-104 | 28 | 426 | 21 | 413 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| A1DPF0 | 1.40e-102 | 28 | 426 | 22 | 414 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 6.84e-102 | 28 | 426 | 21 | 413 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| B0XMA2 | 9.38e-101 | 28 | 426 | 22 | 414 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.30e-100 | 28 | 426 | 22 | 414 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
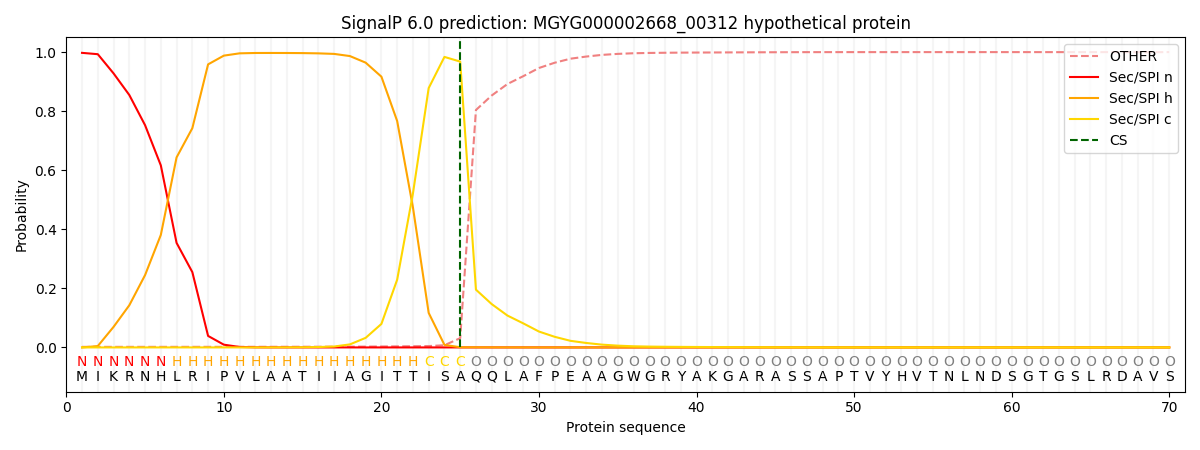
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003399 | 0.994790 | 0.001050 | 0.000279 | 0.000235 | 0.000219 |