You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002685_00303
You are here: Home > Sequence: MGYG000002685_00303
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA9506; ; | |||||||||||
| CAZyme ID | MGYG000002685_00303 | |||||||||||
| CAZy Family | GH156 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6302; End: 9304 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH156 | 490 | 999 | 2.7e-61 | 0.927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00395 | SLH | 1.92e-08 | 285 | 325 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 2.11e-07 | 285 | 440 | 582 | 733 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 6.23e-05 | 344 | 386 | 1 | 42 | S-layer homology domain. |
| cd14791 | GH36 | 1.68e-04 | 633 | 695 | 92 | 158 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| pfam00395 | SLH | 0.002 | 409 | 445 | 1 | 36 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIF01836.1 | 5.48e-123 | 498 | 999 | 49 | 558 |
| QHI69840.1 | 8.16e-105 | 498 | 1000 | 26 | 543 |
| QGJ71964.1 | 6.37e-82 | 466 | 999 | 13 | 550 |
| QHI69841.1 | 9.73e-55 | 498 | 999 | 29 | 538 |
| SMF91957.1 | 2.17e-29 | 279 | 459 | 1051 | 1232 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6RZD_A | 5.26e-12 | 586 | 983 | 104 | 486 | Crystalstructure of an inverting family GH156 exosialidase from uncultured bacterium pG7 [uncultured bacterium pG7],6RZD_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 [uncultured bacterium pG7],6S00_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid [uncultured bacterium pG7],6S00_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid [uncultured bacterium pG7],6S0E_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid [uncultured bacterium pG7],6S0E_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid [uncultured bacterium pG7],6S0F_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid [uncultured bacterium pG7],6S0F_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid [uncultured bacterium pG7] |
| 6BT4_A | 3.46e-10 | 285 | 457 | 26 | 194 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 3.52e-10 | 285 | 457 | 5 | 173 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 6S04_A | 1.02e-09 | 586 | 983 | 104 | 486 | Crystalstructure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid [uncultured bacterium pG7],6S04_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid [uncultured bacterium pG7] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 2.97e-27 | 282 | 463 | 1679 | 1859 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 1.70e-25 | 285 | 462 | 908 | 1084 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P38537 | 8.36e-15 | 285 | 459 | 36 | 206 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P19424 | 1.70e-14 | 292 | 461 | 49 | 216 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P36917 | 4.92e-13 | 285 | 387 | 1056 | 1156 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
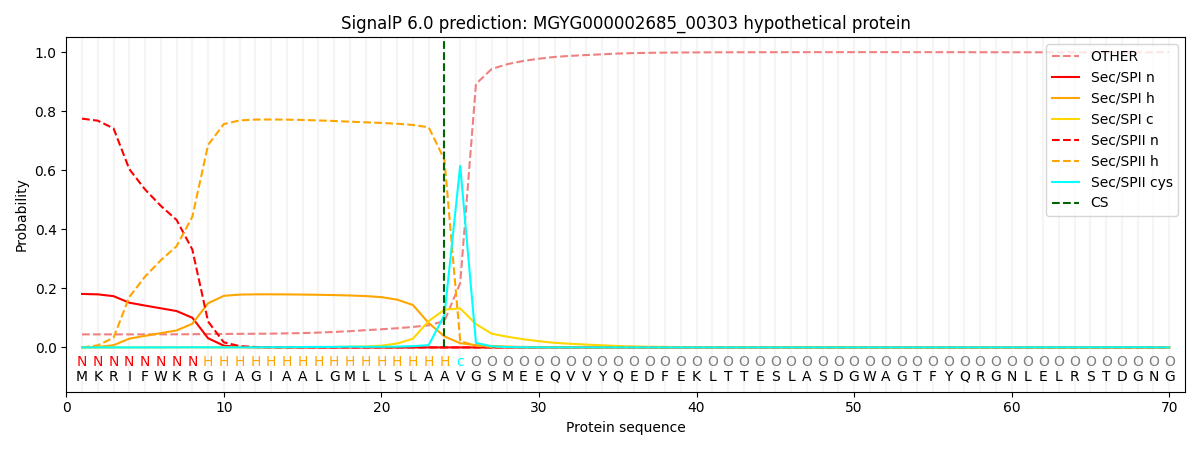
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.045706 | 0.176403 | 0.777456 | 0.000123 | 0.000117 | 0.000183 |
