You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002691_02356
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Dysosmobacter;
|
| CAZyme ID |
MGYG000002691_02356
|
| CAZy Family |
GH79 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002691 |
2941528 |
MAG |
Canada |
North America |
|
| Gene Location |
Start: 8750;
End: 10126
Strand: -
|
No EC number prediction in MGYG000002691_02356.
| Family |
Start |
End |
Evalue |
family coverage |
| GH79 |
78 |
449 |
2.4e-56 |
0.7956043956043956 |
MGYG000002691_02356 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q9FF10
|
6.22e-21 |
81 |
454 |
75 |
438 |
Heparanase-like protein 1 OS=Arabidopsis thaliana OX=3702 GN=At5g07830 PE=2 SV=1 |
|
Q8L608
|
2.05e-15 |
81 |
454 |
72 |
434 |
Heparanase-like protein 2 OS=Arabidopsis thaliana OX=3702 GN=At5g61250 PE=2 SV=1 |
|
Q9LRC8
|
5.97e-13 |
333 |
452 |
326 |
445 |
Baicalin-beta-D-glucuronidase OS=Scutellaria baicalensis OX=65409 GN=SGUS PE=1 SV=1 |
This protein is predicted as OTHER
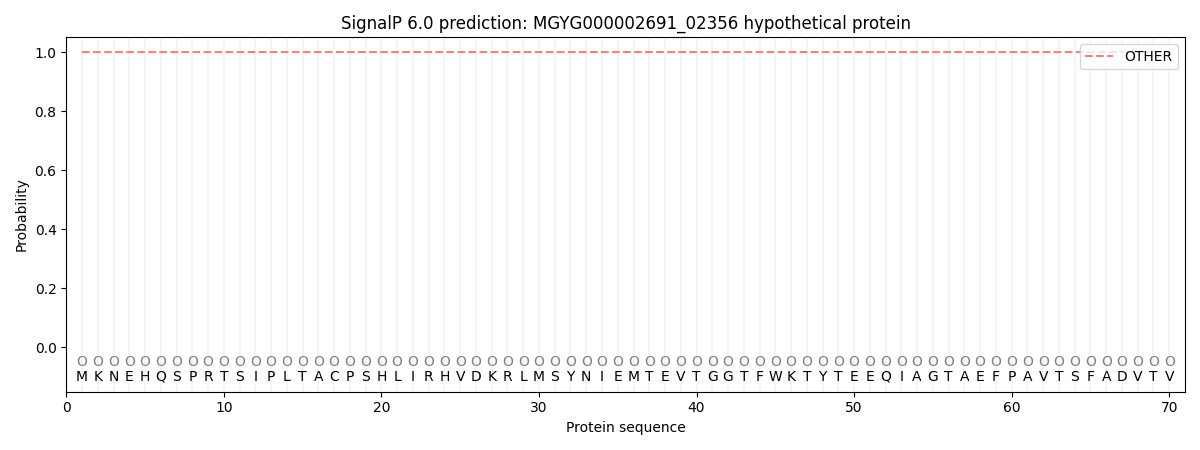
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000021
|
0.000043
|
0.000001
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000002691_02356.

