You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002694_00267
You are here: Home > Sequence: MGYG000002694_00267
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
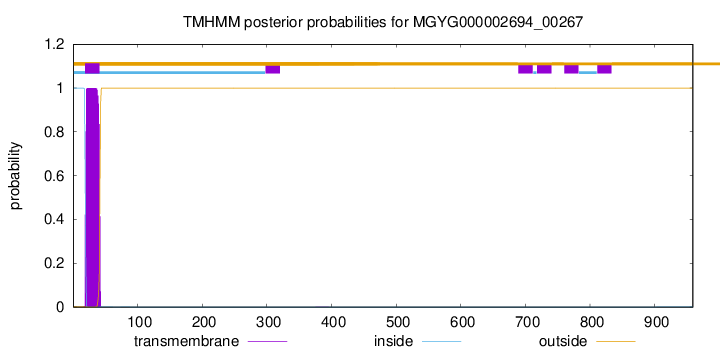
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1591; | |||||||||||
| CAZyme ID | MGYG000002694_00267 | |||||||||||
| CAZy Family | GH97 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2657; End: 5536 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH97 | 56 | 644 | 1e-115 | 0.9793977812995246 |
| CBM51 | 812 | 953 | 1.4e-19 | 0.9552238805970149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14508 | GH97_N | 5.10e-44 | 62 | 300 | 1 | 234 | Glycosyl-hydrolase 97 N-terminal. This N-terminal domain of glycosyl-hydrolase-97 contributes part of the active site pocket. It is also important for contact with the catalytic and C-terminal domains of the whole. |
| pfam10566 | Glyco_hydro_97 | 1.86e-38 | 311 | 549 | 4 | 278 | Glycoside hydrolase 97. This domain is the catalytic region of the bacterial glycosyl-hydrolase family 97. This central part of the GH97 family protein sequences represents a typical and complete (beta/alpha)8-barrel or catalytic TIM-barrel type domain. The N- and C-terminal parts of the sequences, mainly consisting of beta-strands, form two additional non-catalytic domains. In all known glycosidases with the (beta-alpha)8-barrel fold, the amino acid residues at the active site are located on the C-termini of the beta-strands. |
| pfam08305 | NPCBM | 4.13e-20 | 812 | 954 | 5 | 133 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 8.27e-13 | 808 | 954 | 3 | 142 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam13290 | CHB_HEX_C_1 | 3.18e-05 | 739 | 785 | 8 | 55 | Chitobiase/beta-hexosaminidase C-terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXY78499.1 | 5.45e-104 | 58 | 646 | 26 | 627 |
| QNK61828.1 | 8.03e-104 | 53 | 646 | 34 | 629 |
| QNE39615.1 | 3.01e-93 | 50 | 646 | 21 | 620 |
| AGA29066.1 | 3.02e-93 | 58 | 644 | 31 | 631 |
| BCG54503.1 | 2.61e-92 | 59 | 647 | 23 | 627 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XFM_A | 1.34e-40 | 80 | 628 | 40 | 625 | Crystalstructure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_B Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_C Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_D Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron] |
| 3A24_A | 1.91e-35 | 62 | 613 | 6 | 603 | Crystalstructure of BT1871 retaining glycosidase [Bacteroides thetaiotaomicron],3A24_B Crystal structure of BT1871 retaining glycosidase [Bacteroides thetaiotaomicron] |
| 5E1Q_A | 1.60e-34 | 62 | 613 | 20 | 617 | Mutant(D415G) GH97 alpha-galactosidase in complex with Gal-Lac [Bacteroides thetaiotaomicron VPI-5482],5E1Q_B Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D7CFN7 | 8.46e-50 | 61 | 642 | 42 | 618 | Probable retaining alpha-galactosidase OS=Streptomyces bingchenggensis (strain BCW-1) OX=749414 GN=SBI_01652 PE=3 SV=1 |
| Q8A6L0 | 1.66e-35 | 52 | 613 | 17 | 624 | Retaining alpha-galactosidase OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_1871 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
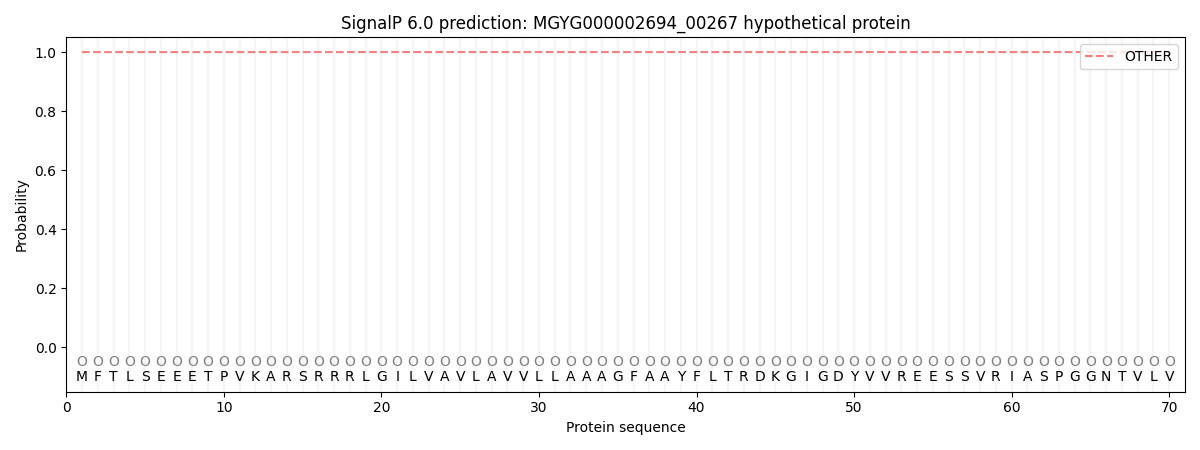
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999997 | 0.000040 | 0.000001 | 0.000000 | 0.000000 | 0.000009 |