You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002694_00978
You are here: Home > Sequence: MGYG000002694_00978
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
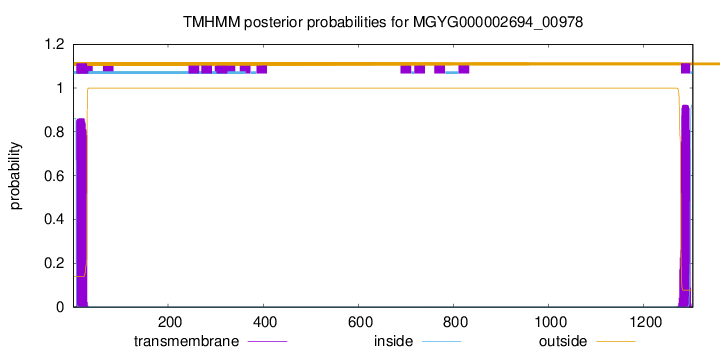
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1591; | |||||||||||
| CAZyme ID | MGYG000002694_00978 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2099; End: 6013 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 317 | 1016 | 1e-62 | 0.7473404255319149 |
| CBM32 | 1024 | 1142 | 2.6e-23 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.40e-22 | 400 | 904 | 113 | 607 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00754 | F5_F8_type_C | 6.22e-17 | 1021 | 1140 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PRK10150 | PRK10150 | 1.20e-10 | 321 | 744 | 14 | 444 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 2.51e-07 | 340 | 475 | 44 | 168 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam02836 | Glyco_hydro_2_C | 3.67e-07 | 665 | 803 | 63 | 210 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNN24101.1 | 1.16e-124 | 317 | 1144 | 40 | 841 |
| ASV74706.1 | 3.63e-120 | 315 | 997 | 32 | 709 |
| QNN24094.1 | 2.46e-107 | 317 | 966 | 52 | 689 |
| ANH81212.1 | 2.30e-98 | 317 | 998 | 28 | 697 |
| QBN20403.1 | 4.34e-96 | 322 | 929 | 32 | 640 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RV9_A | 1.28e-20 | 1016 | 1145 | 8 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 1.32e-20 | 1016 | 1145 | 9 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 2.45e-20 | 1016 | 1145 | 9 | 137 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 1.44e-16 | 1022 | 1145 | 15 | 137 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZZ5_A | 1.48e-16 | 1022 | 1145 | 16 | 138 | X-raycrystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 1.75e-13 | 365 | 721 | 101 | 432 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| T2KPJ7 | 2.41e-10 | 350 | 721 | 81 | 430 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| O52847 | 1.19e-06 | 400 | 725 | 164 | 485 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q8FKG6 | 3.52e-06 | 322 | 768 | 55 | 508 | Beta-galactosidase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
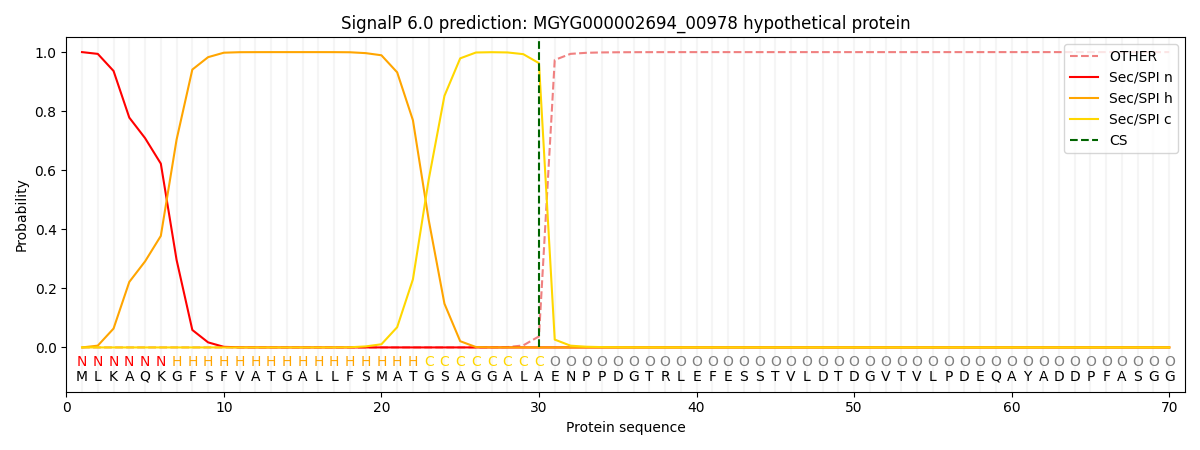
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000328 | 0.998856 | 0.000212 | 0.000248 | 0.000172 | 0.000145 |