You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002702_00426
You are here: Home > Sequence: MGYG000002702_00426
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
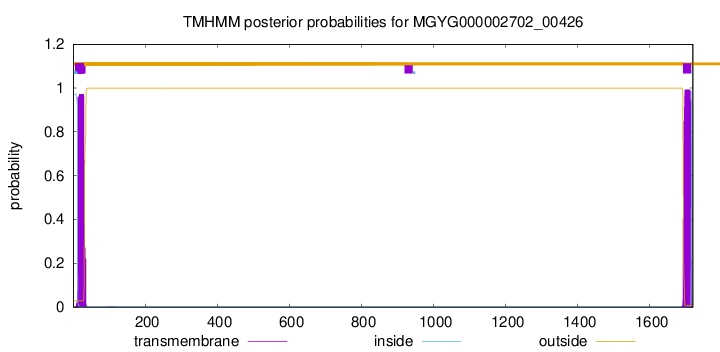
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; ; | |||||||||||
| CAZyme ID | MGYG000002702_00426 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10454; End: 15616 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 432 | 1105 | 2.7e-116 | 0.9957567185289957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12905 | Glyco_hydro_101 | 8.96e-68 | 675 | 940 | 1 | 270 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| cd14244 | GH_101_like | 1.04e-64 | 688 | 966 | 1 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam18080 | Gal_mutarotas_3 | 3.35e-42 | 429 | 674 | 1 | 243 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| pfam17451 | Glyco_hyd_101C | 7.16e-28 | 949 | 1079 | 1 | 111 | Glycosyl hydrolase 101 beta sandwich domain. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by a S. pneumoniae protein, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. This domain represents C-terminal the beta sandwich domain. |
| pfam17974 | GalBD_like | 2.69e-18 | 1266 | 1443 | 1 | 188 | Galactose-binding domain-like. Proteins containing a galactose-binding domain-like fold can be found in several different protein families, in both eukaryotes and prokaryotes. The common function of these domains is to bind to specific ligands, such as cell-surface-attached carbohydrate substrates for galactose oxidase and sialidase, phospholipids on the outer side of the mammalian cell membrane for coagulation factor Va, membrane-anchored ephrin for the Eph family of receptor tyrosine kinases, and a complex of broken single-stranded DNA and DNA polymerase beta for XRCC1. The structure of the galactose-binding domain-like members consists of a beta-sandwich, in which the strands making up the sheets exhibit a jellyroll fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAS61722.1 | 1.35e-186 | 231 | 1647 | 319 | 1728 |
| AYE33559.1 | 1.35e-186 | 231 | 1647 | 319 | 1728 |
| QQA11708.1 | 2.37e-177 | 276 | 1465 | 348 | 1512 |
| AXH51752.1 | 3.27e-177 | 276 | 1452 | 348 | 1496 |
| BAG69287.1 | 4.52e-177 | 276 | 1465 | 348 | 1512 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6QEP_A | 1.29e-84 | 429 | 1434 | 12 | 1091 | EngBFDARPin Fusion 4b H14 [Bifidobacterium longum] |
| 6SH9_B | 1.32e-84 | 429 | 1434 | 12 | 1091 | EngBFDARPin Fusion 4b D12 [Bifidobacterium longum subsp. longum JCM 1217] |
| 6QFK_A | 1.32e-84 | 429 | 1434 | 12 | 1091 | EngBFDARPin Fusion 4b G10 [Bifidobacterium longum] |
| 6QEV_B | 1.32e-84 | 429 | 1434 | 12 | 1091 | EngBFDARPin Fusion 4b B6 [Bifidobacterium longum] |
| 2ZXQ_A | 1.50e-84 | 429 | 1434 | 27 | 1106 | Crystalstructure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 6.06e-95 | 422 | 1448 | 42 | 1037 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q8DR60 | 6.43e-80 | 418 | 1519 | 323 | 1485 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
| Q2MGH6 | 1.47e-79 | 418 | 1519 | 323 | 1485 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| E8MGH9 | 7.76e-12 | 1457 | 1651 | 1671 | 1881 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
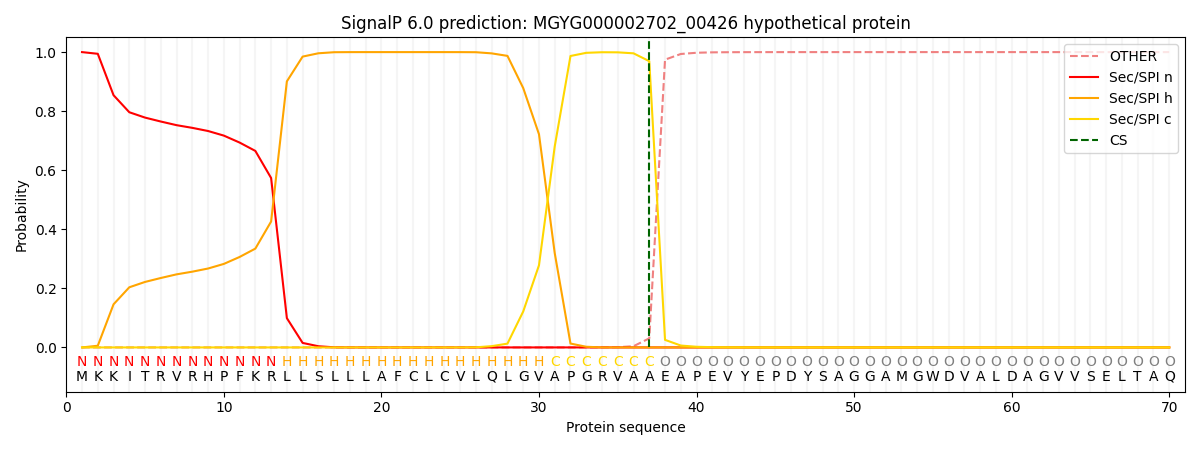
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000362 | 0.998860 | 0.000256 | 0.000188 | 0.000168 | 0.000152 |