You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002709_00477
You are here: Home > Sequence: MGYG000002709_00477
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
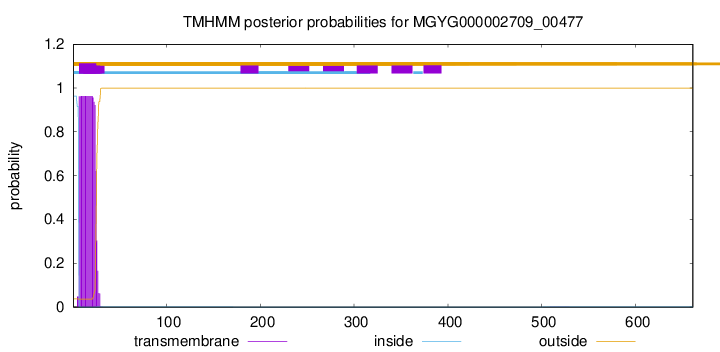
TMHMM annotations
Basic Information help
| Species | UMGS1623 sp900553525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1623; UMGS1623 sp900553525 | |||||||||||
| CAZyme ID | MGYG000002709_00477 | |||||||||||
| CAZy Family | CE2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34872; End: 36857 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 461 | 655 | 8.8e-40 | 0.9952153110047847 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01831 | Endoglucanase_E_like | 1.51e-23 | 461 | 656 | 1 | 169 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| pfam13472 | Lipase_GDSL_2 | 1.58e-12 | 464 | 644 | 1 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 6.99e-11 | 462 | 653 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| COG2755 | TesA | 1.16e-06 | 458 | 658 | 7 | 211 | Lysophospholipase L1 or related esterase [Amino acid transport and metabolism]. |
| cd01822 | Lysophospholipase_L1_like | 9.37e-05 | 460 | 587 | 1 | 101 | Lysophospholipase L1-like subgroup of SGNH-hydrolases. The best characterized member in this family is TesA, an E. coli periplasmic protein with thioesterase, esterase, arylesterase, protease and lysophospholipase activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADC90443.1 | 9.10e-32 | 347 | 658 | 61 | 389 |
| AXB29272.1 | 2.54e-19 | 333 | 659 | 5 | 370 |
| QZN77000.1 | 6.46e-19 | 334 | 660 | 2 | 374 |
| CBL01978.1 | 8.27e-19 | 361 | 659 | 32 | 370 |
| QNU68524.1 | 1.24e-18 | 269 | 661 | 45 | 440 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3U37_A | 2.62e-19 | 421 | 661 | 109 | 406 | AnAcetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_B An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_C An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_D An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_E An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_F An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_G An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],3U37_H An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 2WAO_A | 5.44e-19 | 415 | 661 | 76 | 333 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 2WAB_A | 1.32e-18 | 415 | 661 | 76 | 333 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 4DEV_A | 2.00e-18 | 421 | 661 | 109 | 406 | AnAcetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_B An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_C An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_D An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_E An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_F An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_G An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316],4DEV_H An Acetyl Xylan Esterase (Est2A) from the Rumen Bacterium Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 4XVH_A | 8.30e-14 | 415 | 661 | 64 | 329 | Crystalstructure of a Corynascus thermopiles (Myceliophthora fergusii) carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD) [Chaetomium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10477 | 2.24e-17 | 415 | 661 | 557 | 814 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| B3PDE5 | 3.39e-10 | 355 | 566 | 54 | 250 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ce2C PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
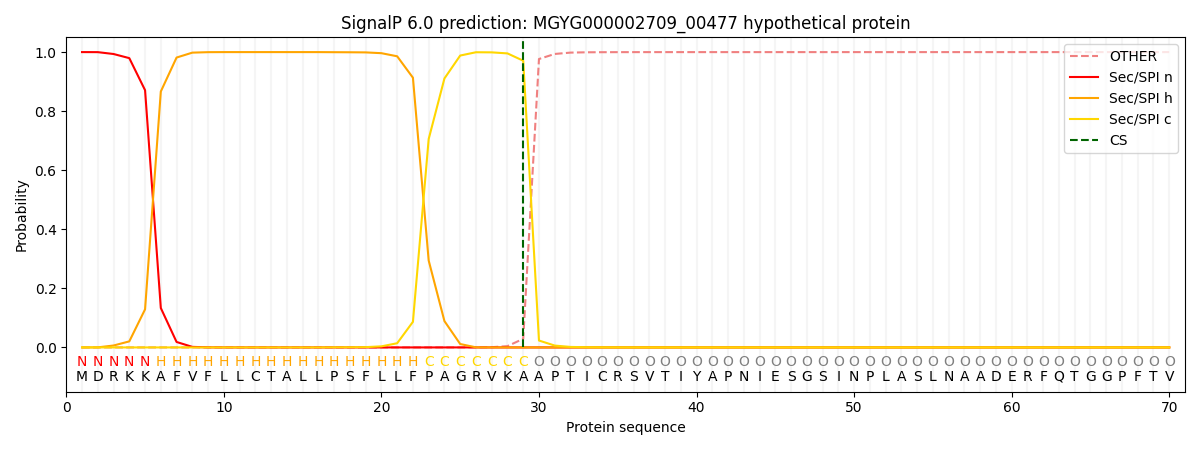
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000263 | 0.999108 | 0.000178 | 0.000173 | 0.000144 | 0.000126 |