You are browsing environment: HUMAN GUT
MGYG000002717_02147
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides;
CAZyme ID
MGYG000002717_02147
CAZy Family
GH2
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002717
5602088
MAG
Canada
North America
Gene Location
Start: 6993;
End: 8072
Strand: -
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam02929
Bgal_small_N
0.005
105
301
2
177
Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase.
more
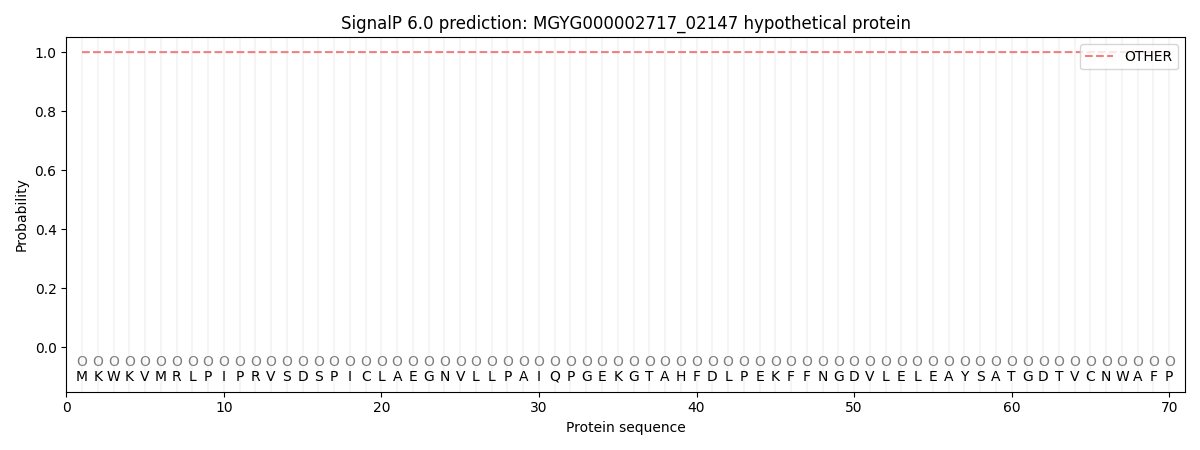
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999926
0.000125
0.000001
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000002717_02147.