You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002719_00416
You are here: Home > Sequence: MGYG000002719_00416
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
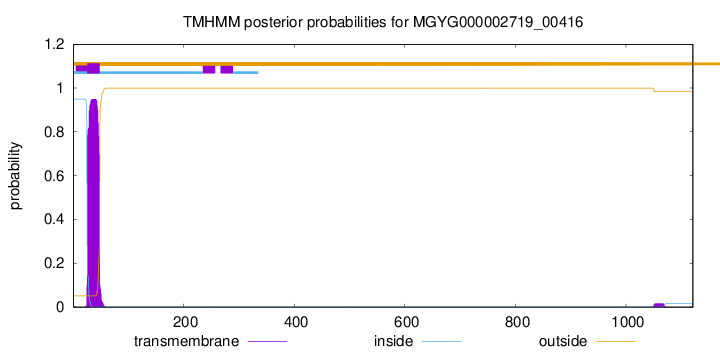
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; HGM11514; HGM11514; ; | |||||||||||
| CAZyme ID | MGYG000002719_00416 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2501; End: 5866 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 552 | 760 | 1.4e-17 | 0.3700189753320683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 5.76e-13 | 552 | 757 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 5.61e-11 | 536 | 671 | 33 | 153 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 2.02e-10 | 615 | 794 | 28 | 178 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam13229 | Beta_helix | 1.11e-08 | 536 | 671 | 10 | 129 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.27e-08 | 614 | 795 | 8 | 141 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE97817.1 | 3.16e-114 | 57 | 1020 | 442 | 1199 |
| QGQ94441.1 | 3.54e-96 | 53 | 1007 | 36 | 802 |
| QJD86952.1 | 1.32e-86 | 56 | 925 | 7 | 704 |
| QGQ94444.1 | 1.13e-85 | 51 | 1007 | 45 | 799 |
| QNK55672.1 | 4.00e-83 | 56 | 922 | 5 | 693 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
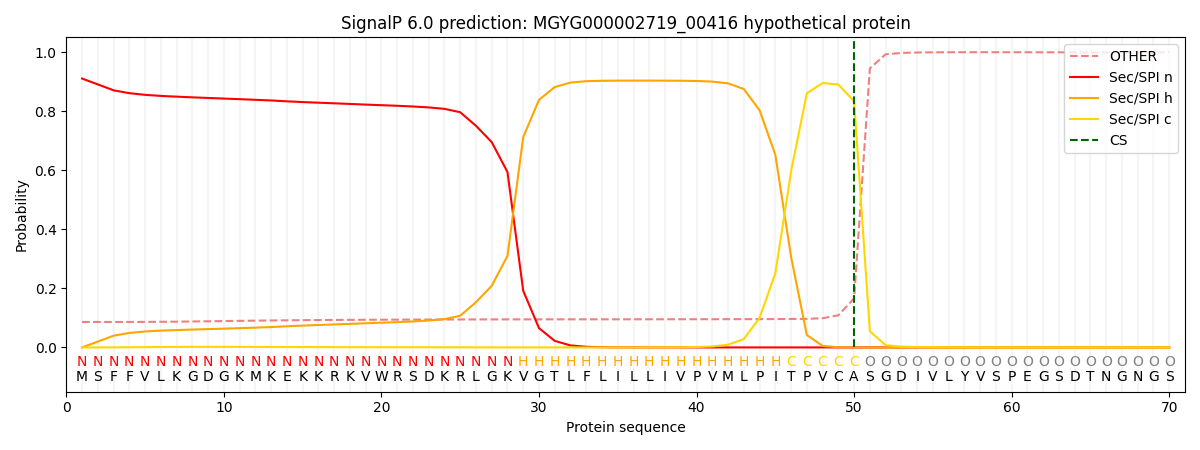
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.093471 | 0.899928 | 0.005312 | 0.000536 | 0.000330 | 0.000401 |