You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002762_00408
You are here: Home > Sequence: MGYG000002762_00408
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900554845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900554845 | |||||||||||
| CAZyme ID | MGYG000002762_00408 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27116; End: 28696 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 4.07e-20 | 13 | 437 | 10 | 441 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.53e-06 | 147 | 374 | 142 | 379 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTO25703.1 | 3.23e-120 | 21 | 519 | 11 | 505 |
| QLK84536.1 | 3.23e-120 | 21 | 519 | 11 | 505 |
| QKH86812.1 | 4.56e-120 | 21 | 519 | 11 | 505 |
| QRP88934.1 | 4.56e-117 | 21 | 503 | 11 | 489 |
| QUU03965.1 | 4.56e-117 | 21 | 503 | 11 | 489 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O67270 | 2.88e-10 | 20 | 399 | 5 | 386 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| Q4K884 | 3.78e-08 | 37 | 339 | 64 | 365 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT2 PE=3 SV=1 |
| B7LM74 | 1.47e-07 | 20 | 291 | 6 | 286 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia fergusonii (strain ATCC 35469 / DSM 13698 / CCUG 18766 / IAM 14443 / JCM 21226 / LMG 7866 / NBRC 102419 / NCTC 12128 / CDC 0568-73) OX=585054 GN=arnT PE=3 SV=2 |
| Q1CIH9 | 1.36e-06 | 5 | 336 | 3 | 332 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Nepal516) OX=377628 GN=arnT PE=3 SV=1 |
| Q1C744 | 1.36e-06 | 5 | 336 | 3 | 332 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
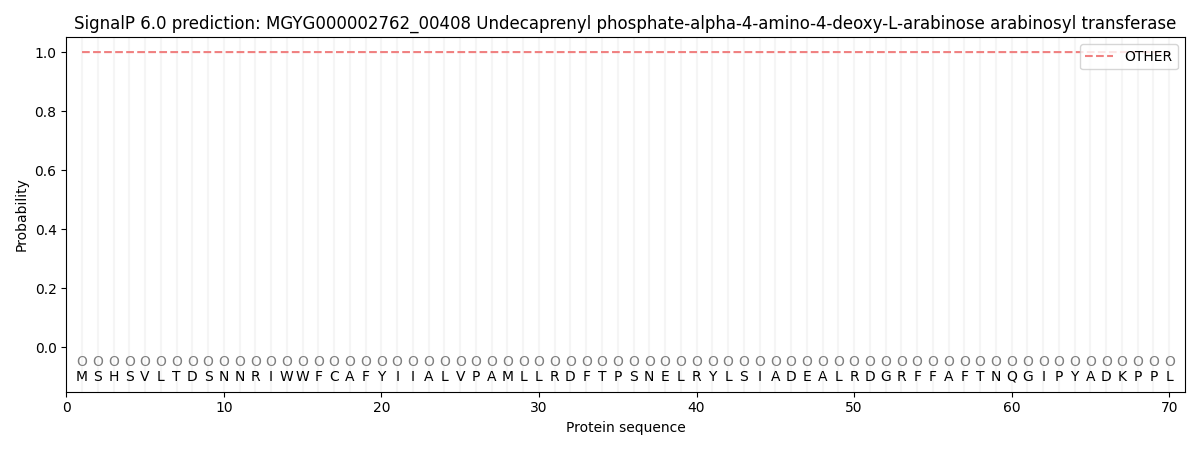
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000038 | 0.000010 | 0.000000 | 0.000000 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
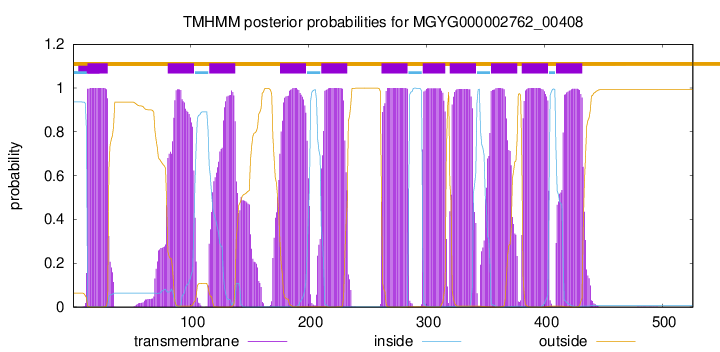
| start | end |
|---|---|
| 13 | 30 |
| 81 | 103 |
| 116 | 138 |
| 176 | 198 |
| 211 | 233 |
| 262 | 284 |
| 297 | 316 |
| 320 | 342 |
| 355 | 377 |
| 381 | 403 |
| 410 | 432 |
