You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002774_01636
You are here: Home > Sequence: MGYG000002774_01636
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Schleiferilactobacillus harbinensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Schleiferilactobacillus; Schleiferilactobacillus harbinensis | |||||||||||
| CAZyme ID | MGYG000002774_01636 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 85738; End: 87981 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 16 | 235 | 1.5e-29 | 0.3962962962962963 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.59e-34 | 14 | 708 | 1 | 523 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.32e-30 | 76 | 229 | 1 | 154 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1928 | PMT1 | 2.37e-06 | 22 | 230 | 26 | 249 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| PRK13279 | arnT | 0.004 | 77 | 238 | 62 | 227 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| PRK12438 | PRK12438 | 0.008 | 297 | 356 | 896 | 945 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHU89858.1 | 5.96e-82 | 12 | 711 | 264 | 889 |
| QHU90800.1 | 5.75e-75 | 12 | 711 | 264 | 886 |
| QJU05422.1 | 7.36e-73 | 12 | 711 | 264 | 890 |
| QUB37647.1 | 2.66e-72 | 26 | 711 | 210 | 780 |
| QLF52252.1 | 1.31e-71 | 12 | 711 | 264 | 891 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P37483 | 5.05e-170 | 17 | 722 | 4 | 662 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
| O34575 | 8.24e-165 | 12 | 720 | 1 | 673 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
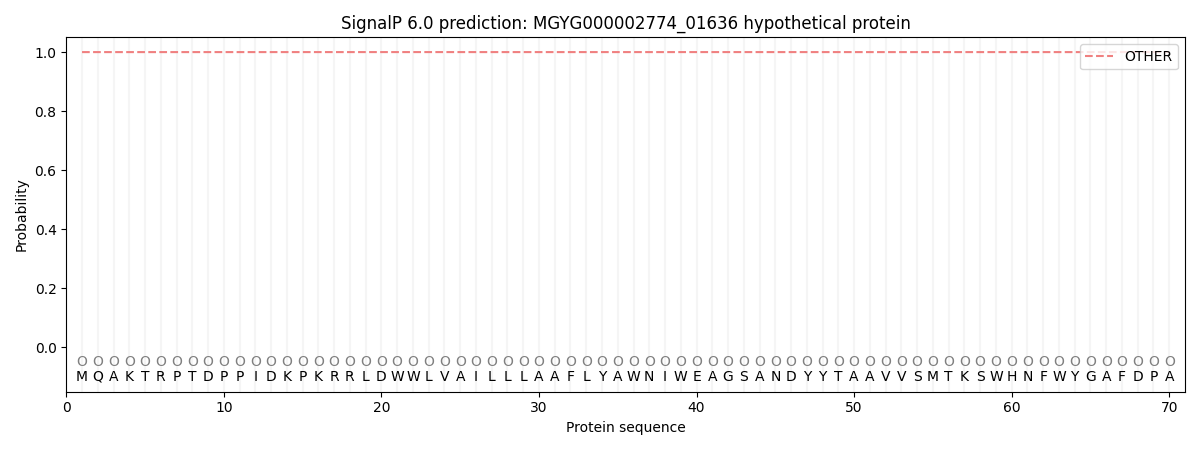
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000026 | 0.000005 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
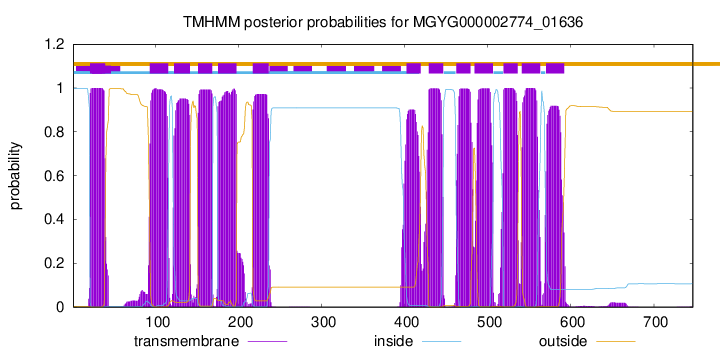
| start | end |
|---|---|
| 21 | 39 |
| 93 | 115 |
| 122 | 141 |
| 151 | 168 |
| 175 | 197 |
| 217 | 236 |
| 402 | 419 |
| 429 | 446 |
| 462 | 479 |
| 484 | 506 |
| 519 | 536 |
| 541 | 563 |
| 570 | 592 |
