You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002781_00248
You are here: Home > Sequence: MGYG000002781_00248
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS775 sp900545325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UMGS775; UMGS775 sp900545325 | |||||||||||
| CAZyme ID | MGYG000002781_00248 | |||||||||||
| CAZy Family | CBM38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30400; End: 32280 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM38 | 255 | 400 | 9.3e-26 | 0.9767441860465116 |
| CBM38 | 35 | 207 | 2e-24 | 0.9767441860465116 |
| CBM38 | 450 | 594 | 1.3e-21 | 0.9767441860465116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033675 | NTTRR-F1 | 7.02e-04 | 33 | 66 | 6 | 31 | NTTRR-F1 domain. NTTRR-F1 (N-terminal To Repetitive Region - Firmicutes 1) is a homology domain found strictly as the N-terminal non-repetitive region of otherwise highly repetitive proteins of various Firmicutes. The repetitive region that follows typically is collagen-like, with every third residue a glycine. |
| NF033675 | NTTRR-F1 | 0.007 | 250 | 282 | 3 | 25 | NTTRR-F1 domain. NTTRR-F1 (N-terminal To Repetitive Region - Firmicutes 1) is a homology domain found strictly as the N-terminal non-repetitive region of otherwise highly repetitive proteins of various Firmicutes. The repetitive region that follows typically is collagen-like, with every third residue a glycine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD82060.1 | 1.17e-91 | 32 | 625 | 58 | 603 |
| AFH61270.2 | 2.94e-91 | 32 | 625 | 83 | 628 |
| AEI40445.1 | 1.82e-90 | 32 | 625 | 83 | 628 |
| BAA24360.1 | 3.87e-75 | 252 | 612 | 59 | 409 |
| CQR54046.1 | 5.98e-70 | 253 | 612 | 75 | 424 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
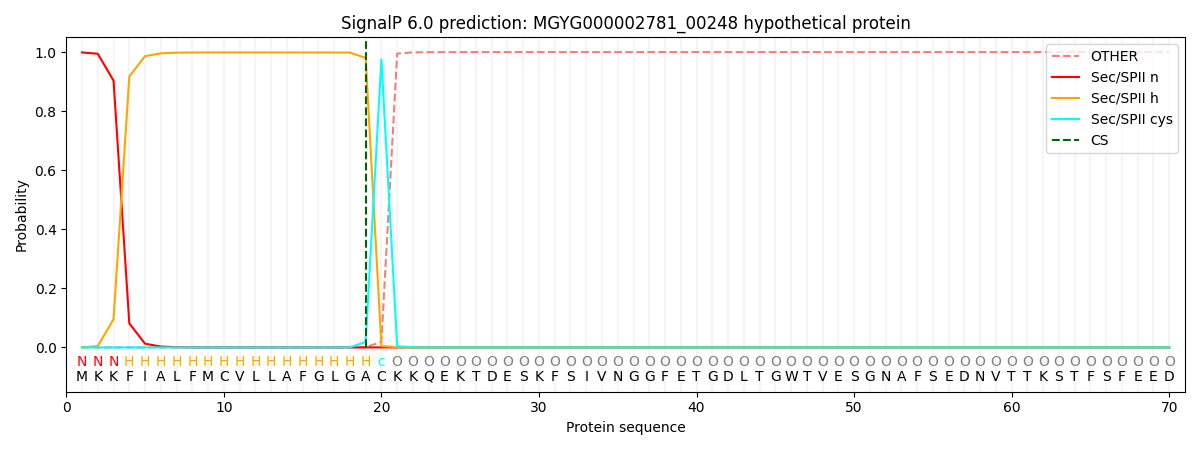
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000003 | 0.001067 | 0.998950 | 0.000000 | 0.000001 | 0.000001 |
