You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002782_00792
You are here: Home > Sequence: MGYG000002782_00792
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Olsenella_E sp900540955 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Atopobiaceae; Olsenella_E; Olsenella_E sp900540955 | |||||||||||
| CAZyme ID | MGYG000002782_00792 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35762; End: 38257 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 1.26e-22 | 202 | 518 | 81 | 347 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.99e-08 | 205 | 471 | 89 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK15098 | PRK15098 | 9.64e-07 | 210 | 514 | 126 | 400 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 2.73e-05 | 509 | 801 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOY60939.1 | 0.0 | 3 | 826 | 5 | 812 |
| BCJ97597.1 | 3.39e-199 | 43 | 823 | 70 | 844 |
| ADK68813.1 | 3.05e-147 | 2 | 822 | 3 | 819 |
| QIA06569.1 | 9.39e-146 | 42 | 819 | 36 | 828 |
| QGY45729.1 | 1.20e-144 | 41 | 819 | 35 | 833 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 4.70e-10 | 97 | 802 | 30 | 669 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 5YOT_A | 1.70e-06 | 175 | 519 | 81 | 399 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 7EAP_A | 1.70e-06 | 175 | 519 | 81 | 399 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 3.42e-35 | 81 | 514 | 36 | 486 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q2UFP8 | 1.28e-30 | 97 | 811 | 40 | 636 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 2.64e-30 | 100 | 801 | 35 | 615 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| B8NGU6 | 1.22e-29 | 162 | 811 | 107 | 632 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
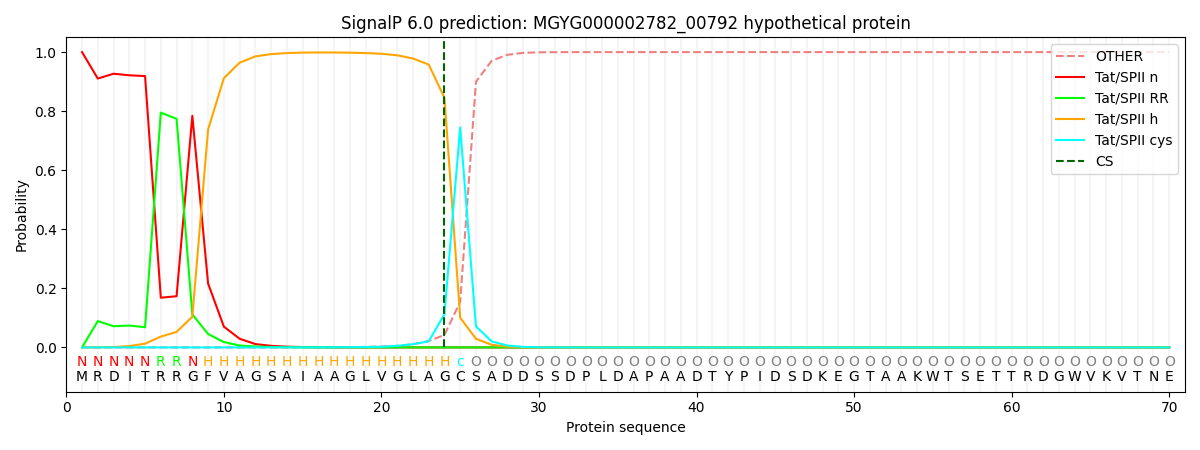
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000017 | 0.000021 | 0.999976 | 0.000000 |
