You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002785_01361
You are here: Home > Sequence: MGYG000002785_01361
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; | |||||||||||
| CAZyme ID | MGYG000002785_01361 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 958; End: 2301 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 174 | 345 | 2.2e-23 | 0.6079295154185022 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 1.35e-59 | 14 | 439 | 6 | 386 | Predicted peptidase [General function prediction only]. |
| pfam18435 | EstA_Ig_like | 2.20e-21 | 34 | 145 | 1 | 119 | Esterase Ig-like N-terminal domain. This is an N-terminal immunoglobulin (Ig)-like domain found in esterases such as EstA. Analysis of the EstA structure confirms that it is a member of the alpha/beta hydrolase family, with a conserved Ser-Asp-His catalytic triad. The Ig-like domain presumably plays a role in the multimerization of EstA into an unusual hexameric structure. Additionally, it may also participate in the catalysis of EstA by guiding the substrate to the active site. |
| COG3509 | LpqC | 1.07e-10 | 173 | 321 | 43 | 177 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| TIGR01840 | esterase_phb | 1.98e-07 | 197 | 321 | 14 | 128 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| pfam10503 | Esterase_phd | 4.54e-06 | 177 | 327 | 1 | 135 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJW36208.1 | 2.36e-41 | 45 | 441 | 693 | 1089 |
| ABS60377.1 | 6.96e-36 | 145 | 443 | 1 | 248 |
| BCI61582.1 | 9.01e-34 | 147 | 441 | 790 | 1044 |
| VTR91196.1 | 4.97e-16 | 176 | 367 | 42 | 206 |
| QEY17854.1 | 9.28e-16 | 177 | 368 | 797 | 994 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 2.84e-58 | 30 | 440 | 4 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 2.75e-12 | 174 | 351 | 18 | 168 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 4Q82_A | 1.91e-09 | 182 | 339 | 70 | 211 | CrystalStructure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365],4Q82_B Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
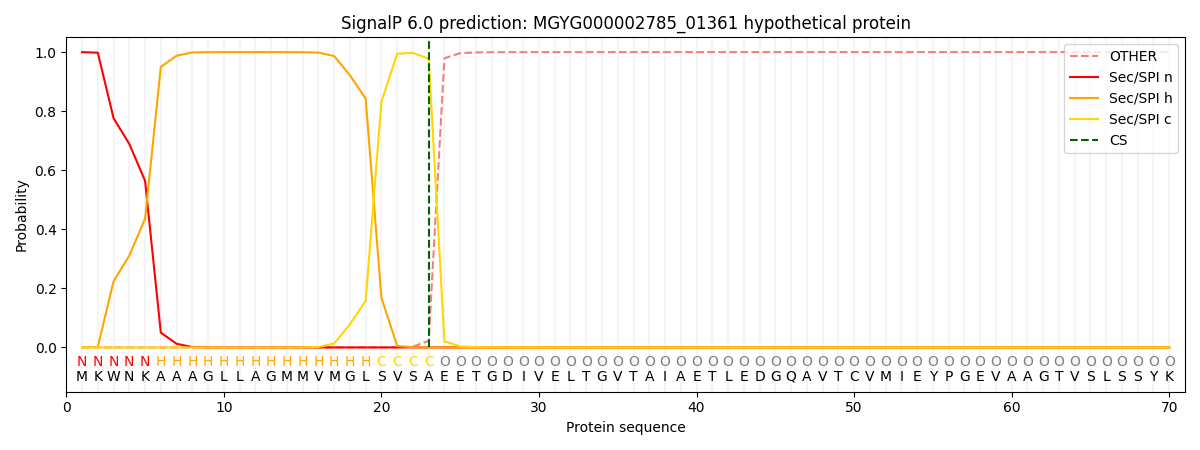
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999057 | 0.000190 | 0.000160 | 0.000151 | 0.000148 |
