You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002827_00503
You are here: Home > Sequence: MGYG000002827_00503
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phyllobacterium sp900539805 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales_A; Rhizobiaceae_A; Phyllobacterium; Phyllobacterium sp900539805 | |||||||||||
| CAZyme ID | MGYG000002827_00503 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10840; End: 12684 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 240 | 464 | 3.4e-34 | 0.9739130434782609 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06427 | CESA_like_2 | 1.33e-132 | 238 | 478 | 1 | 241 | CESA_like_2 is a member of the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, Glucan Biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of glucan. |
| COG1215 | BcsA | 1.06e-45 | 201 | 601 | 20 | 425 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06423 | CESA_like | 4.09e-34 | 242 | 424 | 1 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| pfam13641 | Glyco_tranf_2_3 | 8.41e-30 | 238 | 463 | 2 | 227 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| pfam13632 | Glyco_trans_2_3 | 4.35e-24 | 324 | 515 | 1 | 193 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QND51855.1 | 1.48e-267 | 1 | 605 | 1 | 605 |
| ATU92795.1 | 5.78e-239 | 4 | 605 | 4 | 605 |
| SCD22693.1 | 4.18e-163 | 5 | 602 | 9 | 604 |
| QGA57309.1 | 5.91e-163 | 5 | 602 | 9 | 604 |
| QTN98220.1 | 5.91e-163 | 5 | 602 | 9 | 604 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EJ1_A | 4.02e-08 | 224 | 474 | 115 | 382 | ChainA, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
| 4HG6_A | 4.18e-08 | 224 | 474 | 127 | 394 | ChainA, Cellulose Synthase Subunit A [Cereibacter sphaeroides] |
| 4P00_A | 4.18e-08 | 224 | 474 | 128 | 395 | ChainA, Cellulose Synthase A subunit [Cereibacter sphaeroides 2.4.1],4P02_A Chain A, Cellulose Synthase subunit A [Cereibacter sphaeroides 2.4.1],5EIY_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1],5EJZ_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96587 | 5.79e-19 | 233 | 466 | 44 | 279 | Uncharacterized glycosyltransferase YdaM OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaM PE=3 SV=1 |
| Q8LIY0 | 1.60e-13 | 187 | 469 | 152 | 471 | Probable xyloglucan glycosyltransferase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=CSLC1 PE=3 SV=1 |
| Q9ZQB9 | 8.53e-13 | 233 | 469 | 236 | 479 | Probable xyloglucan glycosyltransferase 12 OS=Arabidopsis thaliana OX=3702 GN=CSLC12 PE=1 SV=1 |
| P74165 | 1.57e-10 | 213 | 472 | 81 | 341 | Beta-monoglucosyldiacylglycerol synthase OS=Synechocystis sp. (strain PCC 6803 / Kazusa) OX=1111708 GN=sll1377 PE=1 SV=1 |
| Q9LR87 | 1.18e-08 | 238 | 466 | 122 | 357 | Probable glucomannan 4-beta-mannosyltransferase 10 OS=Arabidopsis thaliana OX=3702 GN=CSLA10 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
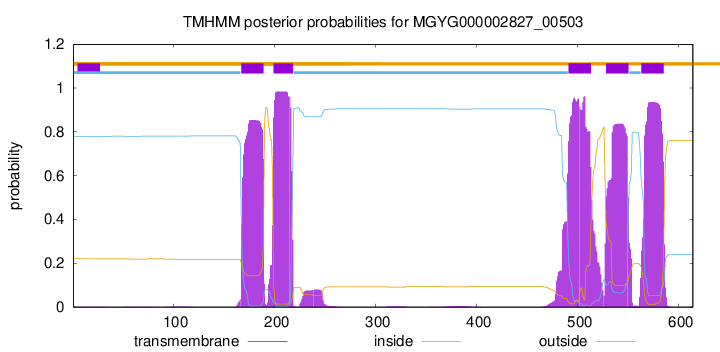
| start | end |
|---|---|
| 167 | 189 |
| 199 | 218 |
| 491 | 513 |
| 528 | 550 |
| 563 | 585 |
