You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002827_02161
You are here: Home > Sequence: MGYG000002827_02161
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phyllobacterium sp900539805 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales_A; Rhizobiaceae_A; Phyllobacterium; Phyllobacterium sp900539805 | |||||||||||
| CAZyme ID | MGYG000002827_02161 | |||||||||||
| CAZy Family | GH103 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66585; End: 67406 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH103 | 37 | 239 | 3.3e-70 | 0.6915254237288135 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02283 | MltB_2 | 4.93e-95 | 36 | 272 | 1 | 300 | lytic murein transglycosylase. Members of this family are closely related to the MltB family lytic murein transglycosylases described by TIGR02282 and are likewise all proteobacterial, although that family and this one form clearly distinct clades. Several species have one member of each family. Many members of this family (unlike the MltB family) contain an additional C-terminal domain, a putative peptidoglycan binding domain (pfam01471), not included in region described by this model. Many sequences appear to contain N-terminal lipoprotein attachment sites, as does E. coli MltB in TIGR02282. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG2951 | MltB | 7.71e-94 | 7 | 273 | 11 | 338 | Membrane-bound lytic murein transglycosylase B [Cell wall/membrane/envelope biogenesis]. |
| pfam13406 | SLT_2 | 2.84e-91 | 36 | 267 | 1 | 291 | Transglycosylase SLT domain. This family is related to the SLT domain pfam01464. |
| TIGR02282 | MltB | 6.37e-36 | 97 | 238 | 58 | 201 | lytic murein transglycosylase B. This family consists of lytic murein transglycosylases (murein hydrolases) in the family of MltB, which is a membrane-bound lipoprotein in Escherichia coli. The N-terminal lipoprotein modification motif is conserved in about half the members of this family. The term Slt35 describes a naturally occurring soluble fragment of MltB. Members of this family never contain the putative peptidoglycan binding domain described by pfam01471, which is associated with several classes of bacterial cell wall lytic enzymes. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| PRK10760 | PRK10760 | 3.88e-24 | 115 | 239 | 143 | 268 | murein hydrolase B; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QND51074.1 | 2.13e-170 | 1 | 273 | 1 | 273 |
| ATU90245.1 | 1.04e-154 | 1 | 273 | 1 | 273 |
| QMV27033.1 | 7.27e-137 | 1 | 273 | 1 | 275 |
| QTN97686.1 | 7.27e-137 | 1 | 273 | 1 | 275 |
| AEK55119.1 | 1.47e-136 | 1 | 273 | 1 | 275 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5ANZ_A | 3.63e-37 | 37 | 237 | 48 | 250 | CrystalStructure of SltB3 from Pseudomonas aeruginosa. [Pseudomonas aeruginosa],5AO7_A Crystal Structure of SltB3 from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa] |
| 5AO8_A | 3.63e-37 | 37 | 237 | 48 | 250 | CrystalStructure of SltB3 from Pseudomonas aeruginosa in complex with NAG-NAM-pentapeptide [Pseudomonas aeruginosa] |
| 5O8X_A | 2.22e-21 | 97 | 237 | 66 | 208 | TheX-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa],5O8X_B The X-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 4ANR_A | 2.80e-21 | 97 | 237 | 83 | 225 | Crystalstructure of soluble lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 1LTM_A | 5.26e-17 | 118 | 237 | 107 | 227 | AcceleratedX-ray Structure Elucidation Of A 36 Kda Muramidase/transglycosylase Using Warp [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P41052 | 4.08e-16 | 118 | 237 | 148 | 268 | Membrane-bound lytic murein transglycosylase B OS=Escherichia coli (strain K12) OX=83333 GN=mltB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
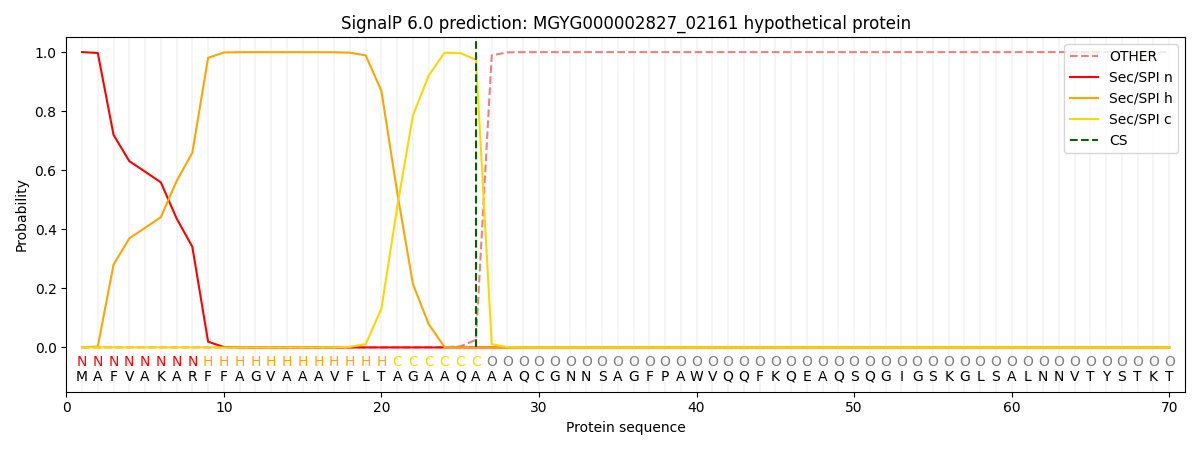
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000351 | 0.998974 | 0.000221 | 0.000171 | 0.000143 | 0.000134 |
