You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002827_03775
Basic Information
help
| Species |
Phyllobacterium sp900539805
|
| Lineage |
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales_A; Rhizobiaceae_A; Phyllobacterium; Phyllobacterium sp900539805
|
| CAZyme ID |
MGYG000002827_03775
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002827 |
5102924 |
MAG |
Singapore |
Asia |
|
| Gene Location |
Start: 37295;
End: 38473
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
39 |
353 |
3.5e-154 |
0.9936507936507937 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
3.10e-15 |
29 |
341 |
13 |
267 |
Cellulase (glycosyl hydrolase family 5). |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
W8QRE4
|
1.69e-06 |
105 |
350 |
125 |
345 |
Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
This protein is predicted as SP
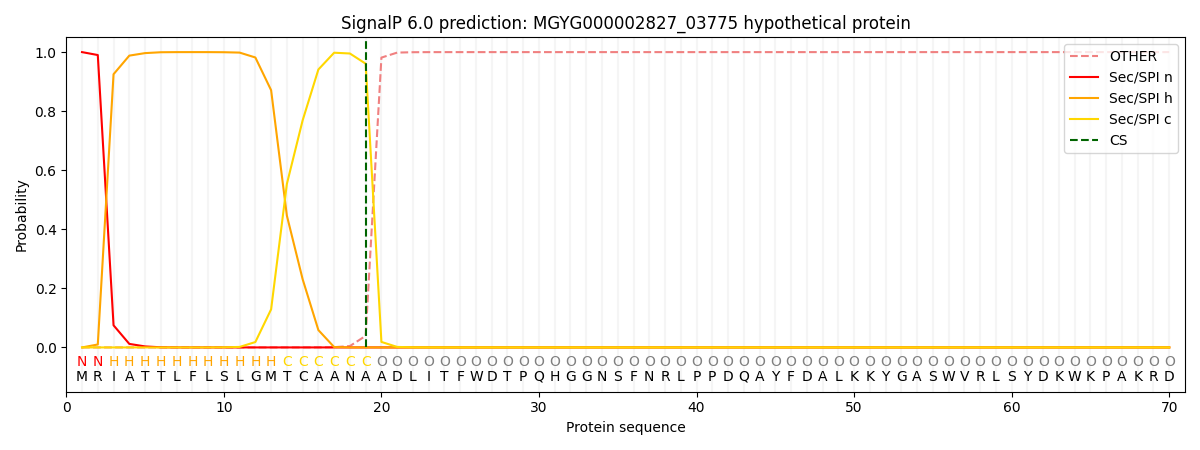
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000333
|
0.998985
|
0.000175
|
0.000179
|
0.000154
|
0.000144
|
There is no transmembrane helices in MGYG000002827_03775.

