You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002834_00427
You are here: Home > Sequence: MGYG000002834_00427
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
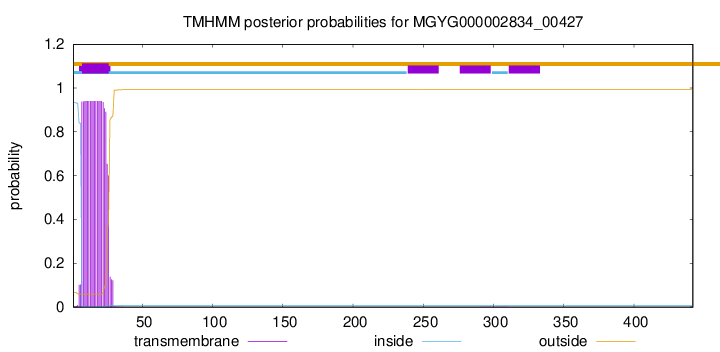
TMHMM annotations
Basic Information help
| Species | Prevotella sp900551985 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900551985 | |||||||||||
| CAZyme ID | MGYG000002834_00427 | |||||||||||
| CAZy Family | CE7 | |||||||||||
| CAZyme Description | Acetyl esterase Axe7A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18540; End: 19868 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE7 | 127 | 435 | 3.9e-78 | 0.9616613418530351 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3458 | Axe1 | 1.91e-39 | 128 | 426 | 13 | 303 | Cephalosporin-C deacetylase or related acetyl esterase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam05448 | AXE1 | 2.17e-32 | 127 | 422 | 8 | 299 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
| COG0412 | DLH | 1.47e-04 | 183 | 321 | 11 | 143 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG2936 | COG2936 | 0.001 | 181 | 310 | 27 | 144 | Predicted acyl esterase [General function prediction only]. |
| pfam10142 | PhoPQ_related | 0.001 | 274 | 423 | 153 | 302 | PhoPQ-activated pathogenicity-related protein. Members of this family of bacterial proteins are involved in the virulence of some pathogenic proteobacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67783.1 | 1.89e-303 | 14 | 442 | 1 | 429 |
| BCS86240.1 | 4.61e-225 | 35 | 441 | 2 | 396 |
| AGH13969.1 | 9.71e-220 | 20 | 440 | 6 | 414 |
| QMI79435.1 | 2.13e-178 | 22 | 440 | 17 | 423 |
| QBJ17988.1 | 2.13e-178 | 22 | 440 | 17 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GMA_A | 1.96e-25 | 127 | 422 | 24 | 315 | Crystalstructure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_B Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_C Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_D Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_E Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_F Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8] |
| 3M81_A | 2.69e-25 | 127 | 422 | 24 | 315 | Crystalstructure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],3M81_B Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],3M81_C Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],3M81_D Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],3M81_E Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],3M81_F Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.50 A resolution (native apo structure) [Thermotoga maritima],5FDF_A Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5FDF_B Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5FDF_C Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5FDF_D Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5FDF_E Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5FDF_F Crystal structure of the monoclinic form of Thermotoga maritima Acetyl Esterase TM0077 (apo structure) at 1.76 Angstrom resolution [Thermotoga maritima],5JIB_A Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8],5JIB_B Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8],5JIB_C Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8],5JIB_D Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8],5JIB_E Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8],5JIB_F Crystal structure of the Thermotoga maritima acetyl esterase (TM0077) complex with a substrate analog [Thermotoga maritima MSB8] |
| 1L7A_A | 6.91e-25 | 127 | 422 | 9 | 298 | structuralGenomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis],1L7A_B structural Genomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis] |
| 1VLQ_A | 9.40e-25 | 127 | 422 | 24 | 315 | Crystalstructure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_B Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_C Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_D Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_E Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_F Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_G Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_H Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_I Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_J Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_K Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8],1VLQ_L Crystal structure of Acetyl xylan esterase (TM0077) from Thermotoga maritima at 2.10 A resolution [Thermotoga maritima MSB8] |
| 3M82_A | 1.29e-24 | 127 | 422 | 24 | 315 | Crystalstructure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M82_B Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M82_C Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M82_D Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M82_E Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M82_F Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.40 A resolution (PMSF inhibitor complex structure) [Thermotoga maritima],3M83_A Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima],3M83_B Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima],3M83_C Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima],3M83_D Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima],3M83_E Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima],3M83_F Crystal structure of Acetyl xylan esterase (TM0077) from THERMOTOGA MARITIMA at 2.12 A resolution (paraoxon inhibitor complex structure) [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXI2 | 2.59e-77 | 37 | 423 | 46 | 421 | Acetyl esterase Axe7A OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe7A PE=1 SV=1 |
| Q9WXT2 | 1.21e-24 | 127 | 422 | 12 | 303 | Cephalosporin-C deacetylase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=axeA PE=1 SV=1 |
| P94388 | 7.09e-24 | 127 | 422 | 9 | 298 | Cephalosporin-C deacetylase OS=Bacillus subtilis (strain 168) OX=224308 GN=cah PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
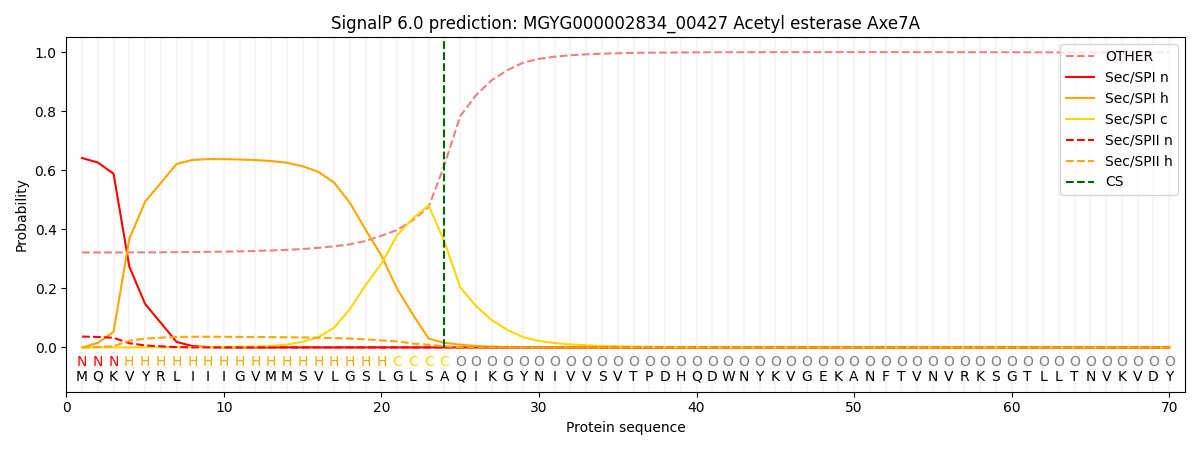
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.334377 | 0.625162 | 0.038550 | 0.000476 | 0.000422 | 0.001000 |