You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002836_00343
You are here: Home > Sequence: MGYG000002836_00343
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
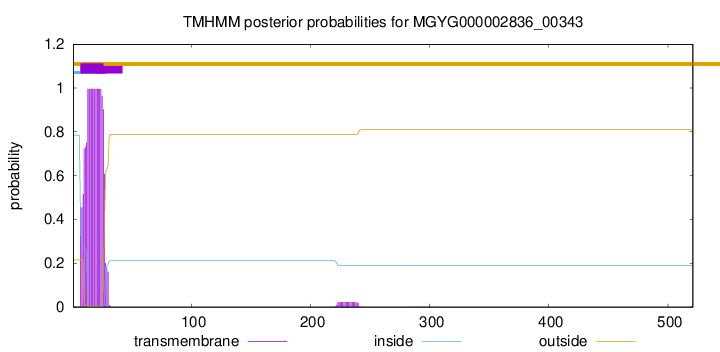
TMHMM annotations
Basic Information help
| Species | Anaerocolumna aminovalerica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerocolumna; Anaerocolumna aminovalerica | |||||||||||
| CAZyme ID | MGYG000002836_00343 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74954; End: 76519 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 228 | 499 | 6.2e-42 | 0.7630769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 4.47e-10 | 243 | 341 | 201 | 315 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 6.45e-09 | 252 | 422 | 72 | 238 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02793 | PLN02793 | 1.08e-06 | 232 | 400 | 140 | 335 | Probable polygalacturonase |
| PLN03010 | PLN03010 | 7.60e-05 | 245 | 465 | 143 | 340 | polygalacturonase |
| PLN03003 | PLN03003 | 0.010 | 245 | 438 | 117 | 298 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCK00431.1 | 5.42e-300 | 1 | 521 | 1 | 520 |
| ABX43664.1 | 6.20e-247 | 16 | 521 | 13 | 518 |
| ACI19833.1 | 2.88e-151 | 6 | 512 | 2 | 476 |
| ACK41503.1 | 5.76e-151 | 16 | 511 | 6 | 475 |
| QTE67472.1 | 2.92e-129 | 48 | 511 | 33 | 478 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 3.79e-06 | 226 | 388 | 153 | 331 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
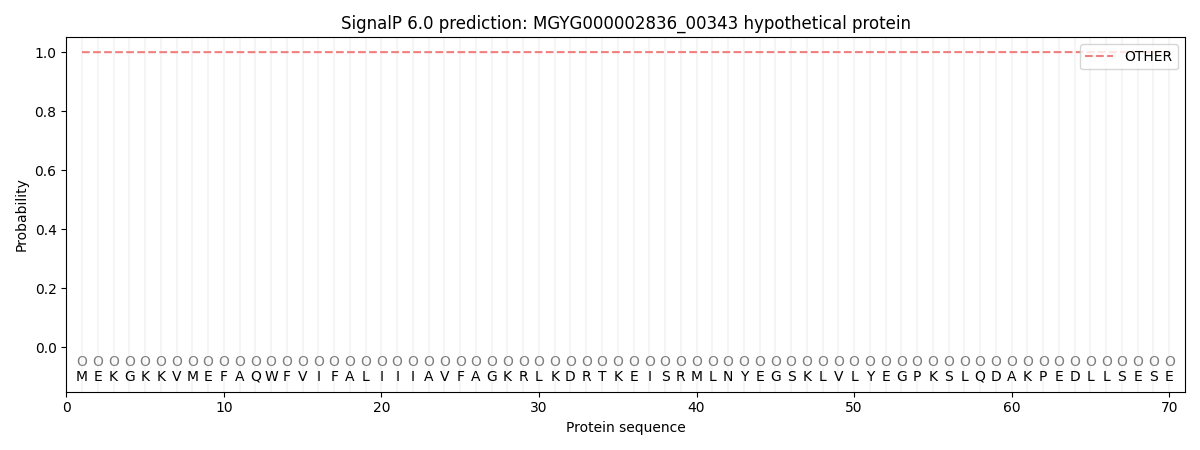
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999763 | 0.000220 | 0.000014 | 0.000001 | 0.000001 | 0.000038 |