You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002836_02325
You are here: Home > Sequence: MGYG000002836_02325
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerocolumna aminovalerica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerocolumna; Anaerocolumna aminovalerica | |||||||||||
| CAZyme ID | MGYG000002836_02325 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 86167; End: 87570 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 39 | 288 | 3.1e-23 | 0.6925675675675675 |
| CBM12 | 423 | 455 | 6.5e-17 | 0.9411764705882353 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02871 | GH18_chitinase_D-like | 6.73e-123 | 36 | 324 | 1 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| COG3469 | Chi1 | 1.38e-42 | 37 | 332 | 27 | 332 | Chitinase [Carbohydrate transport and metabolism]. |
| pfam13290 | CHB_HEX_C_1 | 3.65e-23 | 344 | 408 | 1 | 65 | Chitobiase/beta-hexosaminidase C-terminal domain. |
| pfam00704 | Glyco_hydro_18 | 9.64e-19 | 40 | 310 | 4 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 1.10e-18 | 40 | 311 | 4 | 287 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX42168.1 | 1.82e-161 | 19 | 328 | 247 | 550 |
| BCJ98159.1 | 2.51e-156 | 27 | 328 | 249 | 550 |
| BBH86234.1 | 1.80e-154 | 31 | 328 | 202 | 500 |
| QUQ72090.1 | 4.29e-151 | 33 | 328 | 264 | 558 |
| BBH95642.1 | 1.00e-147 | 33 | 328 | 232 | 528 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EBV_A | 2.12e-135 | 36 | 330 | 5 | 296 | Crystalstructure of putative Chitinase A from Streptomyces coelicolor. [Streptomyces coelicolor] |
| 5KZ6_A | 2.75e-71 | 27 | 328 | 1 | 332 | 1.25Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis],5KZ6_B 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis] |
| 3N11_A | 1.39e-70 | 31 | 328 | 2 | 329 | Crystalstricture of wild-type chitinase from Bacillus cereus NCTU2 [Bacillus cereus],3N12_A Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N15_A | 3.89e-70 | 31 | 328 | 2 | 329 | Crystalstricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N17_A | 1.53e-69 | 31 | 328 | 2 | 329 | Crystalstricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 6.52e-58 | 38 | 328 | 192 | 514 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| Q05638 | 4.22e-47 | 24 | 320 | 252 | 591 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
| D4AVJ0 | 1.28e-46 | 54 | 328 | 17 | 331 | Probable class II chitinase ARB_00204 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00204 PE=1 SV=2 |
| A5FB63 | 7.01e-33 | 33 | 324 | 1139 | 1476 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| E9F7R6 | 4.54e-28 | 11 | 325 | 9 | 345 | Endochitinase 4 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
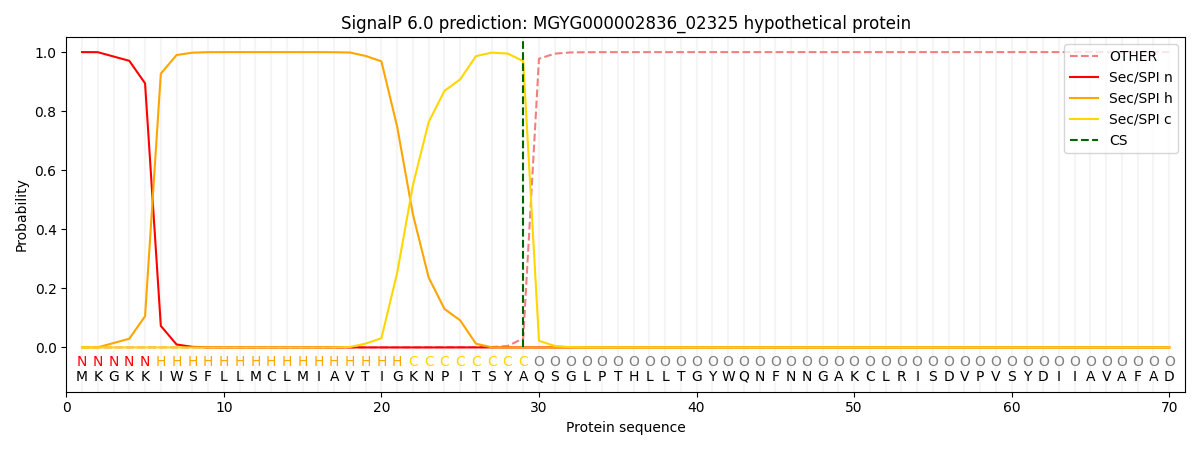
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000302 | 0.998993 | 0.000205 | 0.000169 | 0.000152 | 0.000146 |

