You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002875_00041
You are here: Home > Sequence: MGYG000002875_00041
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900541865 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900541865 | |||||||||||
| CAZyme ID | MGYG000002875_00041 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43279; End: 47118 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 345 | 635 | 4.2e-67 | 0.9826388888888888 |
| PL1 | 784 | 986 | 1.1e-58 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 4.07e-48 | 339 | 635 | 2 | 296 | pectinesterase |
| PLN02432 | PLN02432 | 6.34e-43 | 339 | 635 | 8 | 286 | putative pectinesterase |
| pfam01095 | Pectinesterase | 4.69e-40 | 345 | 635 | 3 | 295 | Pectinesterase. |
| PLN02682 | PLN02682 | 1.57e-38 | 340 | 635 | 67 | 361 | pectinesterase family protein |
| COG4677 | PemB | 6.41e-36 | 342 | 631 | 83 | 398 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU63102.1 | 0.0 | 10 | 1208 | 14 | 1221 |
| ASB38821.1 | 0.0 | 10 | 1208 | 14 | 1221 |
| QQR09559.1 | 0.0 | 10 | 1208 | 14 | 1221 |
| QCP72287.1 | 0.0 | 17 | 1277 | 32 | 1315 |
| QCD38595.1 | 0.0 | 17 | 1277 | 32 | 1315 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C1C_A | 6.43e-28 | 338 | 614 | 5 | 271 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 6.43e-28 | 338 | 614 | 5 | 271 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 1GQ8_A | 4.57e-24 | 340 | 617 | 7 | 281 | Pectinmethylesterase from Carrot [Daucus carota] |
| 3UW0_A | 2.07e-21 | 317 | 591 | 9 | 310 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 2NSP_A | 6.88e-15 | 341 | 635 | 6 | 334 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 1.43e-41 | 727 | 1196 | 19 | 417 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 3.53e-41 | 727 | 1196 | 19 | 417 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| B0XMA2 | 2.99e-40 | 727 | 1193 | 20 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q4WL88 | 4.04e-40 | 727 | 1193 | 20 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 2.41e-39 | 727 | 1196 | 19 | 417 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
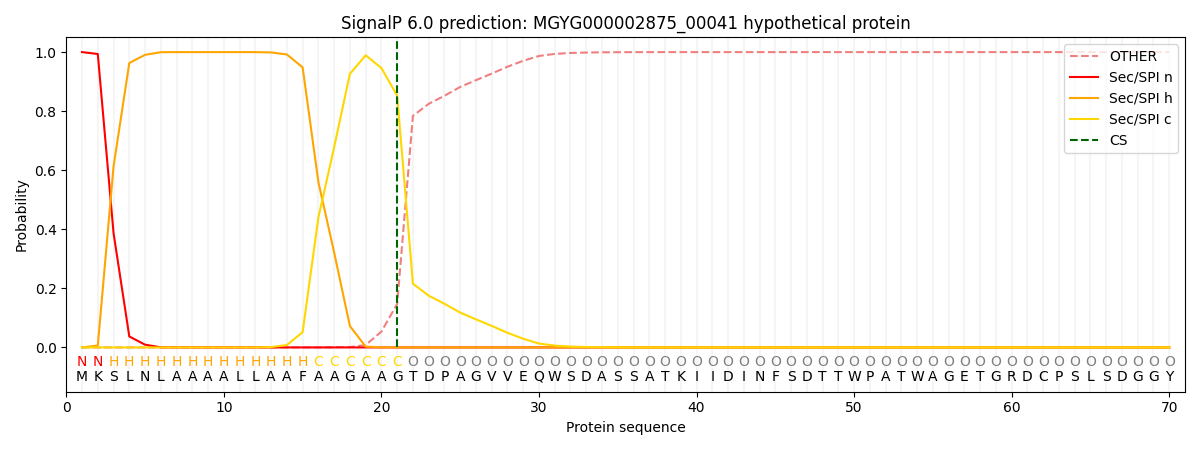
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000265 | 0.999131 | 0.000158 | 0.000168 | 0.000140 | 0.000136 |
