You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002875_00477
You are here: Home > Sequence: MGYG000002875_00477
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900541865 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900541865 | |||||||||||
| CAZyme ID | MGYG000002875_00477 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18318; End: 22445 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 966 | 1236 | 5.8e-42 | 0.9340277777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02497 | PLN02497 | 7.28e-16 | 1040 | 1210 | 116 | 299 | probable pectinesterase |
| PLN02432 | PLN02432 | 1.39e-13 | 1054 | 1244 | 107 | 291 | putative pectinesterase |
| pfam01095 | Pectinesterase | 6.11e-13 | 984 | 1240 | 29 | 297 | Pectinesterase. |
| PLN02773 | PLN02773 | 6.62e-13 | 1091 | 1232 | 152 | 290 | pectinesterase |
| COG4677 | PemB | 3.48e-12 | 934 | 1160 | 60 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37302.1 | 1.25e-253 | 391 | 1375 | 173 | 1159 |
| ANU64595.1 | 1.25e-253 | 391 | 1375 | 173 | 1159 |
| QQR08039.1 | 1.25e-253 | 391 | 1375 | 173 | 1159 |
| QCD38476.1 | 6.47e-67 | 854 | 1373 | 934 | 1473 |
| QCP72166.1 | 6.47e-67 | 854 | 1373 | 934 | 1473 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 4.02e-16 | 986 | 1239 | 36 | 335 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 4PMH_A | 1.07e-06 | 1076 | 1162 | 192 | 294 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4PSN0 | 2.12e-14 | 1031 | 1182 | 108 | 272 | Probable pectinesterase 29 OS=Arabidopsis thaliana OX=3702 GN=PME29 PE=2 SV=1 |
| Q3E9D3 | 1.16e-13 | 1031 | 1182 | 103 | 267 | Probable pectinesterase 55 OS=Arabidopsis thaliana OX=3702 GN=PME55 PE=2 SV=1 |
| Q4PSQ5 | 4.19e-10 | 1035 | 1182 | 112 | 271 | Probable pectinesterase 66 OS=Arabidopsis thaliana OX=3702 GN=PME66 PE=2 SV=2 |
| O64479 | 4.29e-10 | 1013 | 1182 | 96 | 276 | Putative pectinesterase 10 OS=Arabidopsis thaliana OX=3702 GN=PME10 PE=2 SV=1 |
| Q9LUL8 | 3.19e-09 | 986 | 1244 | 690 | 957 | Putative pectinesterase/pectinesterase inhibitor 26 OS=Arabidopsis thaliana OX=3702 GN=PME26 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
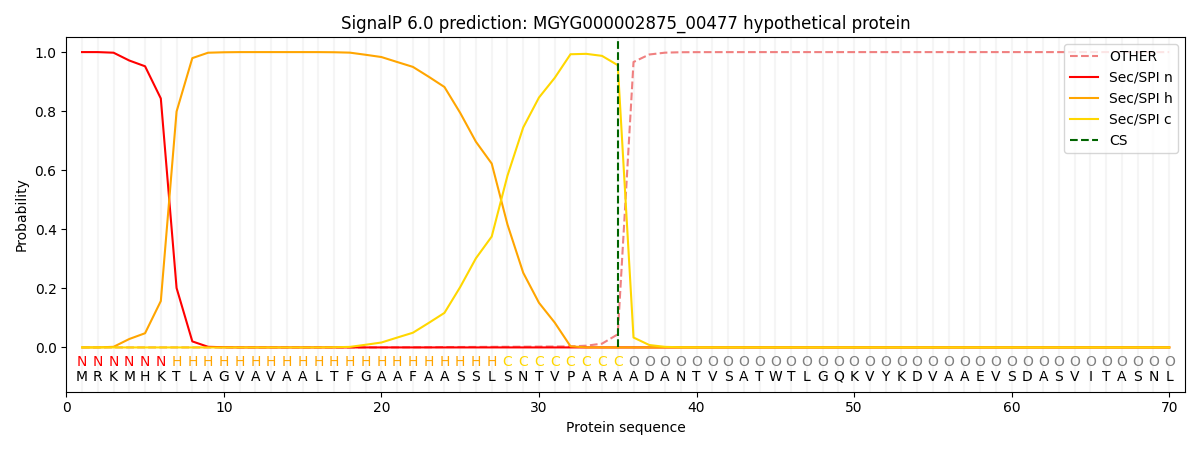
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.999027 | 0.000157 | 0.000203 | 0.000170 | 0.000148 |

