You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002875_01112
You are here: Home > Sequence: MGYG000002875_01112
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900541865 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900541865 | |||||||||||
| CAZyme ID | MGYG000002875_01112 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | Isomalto-dextranase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42213; End: 43634 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 201 | 451 | 4.2e-33 | 0.9344978165938864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17801 | Melibiase_C | 6.03e-21 | 398 | 471 | 1 | 74 | Alpha galactosidase C-terminal beta sandwich domain. This domain is found at the C-terminus of alpha galactosidase enzymes. |
| PLN02229 | PLN02229 | 4.13e-11 | 340 | 468 | 279 | 414 | alpha-galactosidase |
| PLN02808 | PLN02808 | 2.12e-07 | 303 | 472 | 212 | 384 | alpha-galactosidase |
| PLN02692 | PLN02692 | 7.08e-07 | 349 | 468 | 282 | 405 | alpha-galactosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67171.1 | 1.60e-240 | 1 | 471 | 1 | 473 |
| QUU07210.1 | 1.18e-206 | 34 | 473 | 40 | 492 |
| QUT44530.1 | 4.79e-206 | 34 | 473 | 40 | 492 |
| QUT79686.1 | 4.79e-206 | 34 | 473 | 40 | 492 |
| QRM98947.1 | 4.79e-206 | 34 | 473 | 40 | 492 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AWO_A | 3.33e-93 | 49 | 471 | 37 | 465 | Arthrobacterglobiformis T6 isomalto-dextranse [Arthrobacter globiformis],5AWP_A Arthrobacter globiformis T6 isomalto-dextranase complexed with isomaltose [Arthrobacter globiformis],5AWQ_A Arthrobacter globiformis T6 isomalto-dextranse complexed with panose [Arthrobacter globiformis] |
| 1UAS_A | 6.17e-14 | 201 | 468 | 117 | 356 | ChainA, alpha-galactosidase [Oryza sativa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44052 | 2.58e-93 | 49 | 471 | 63 | 491 | Isomalto-dextranase OS=Arthrobacter globiformis OX=1665 GN=imd PE=1 SV=1 |
| Q9FXT4 | 4.62e-13 | 201 | 468 | 172 | 411 | Alpha-galactosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os10g0493600 PE=1 SV=1 |
| Q8VXZ7 | 5.76e-08 | 329 | 466 | 277 | 422 | Alpha-galactosidase 3 OS=Arabidopsis thaliana OX=3702 GN=AGAL3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
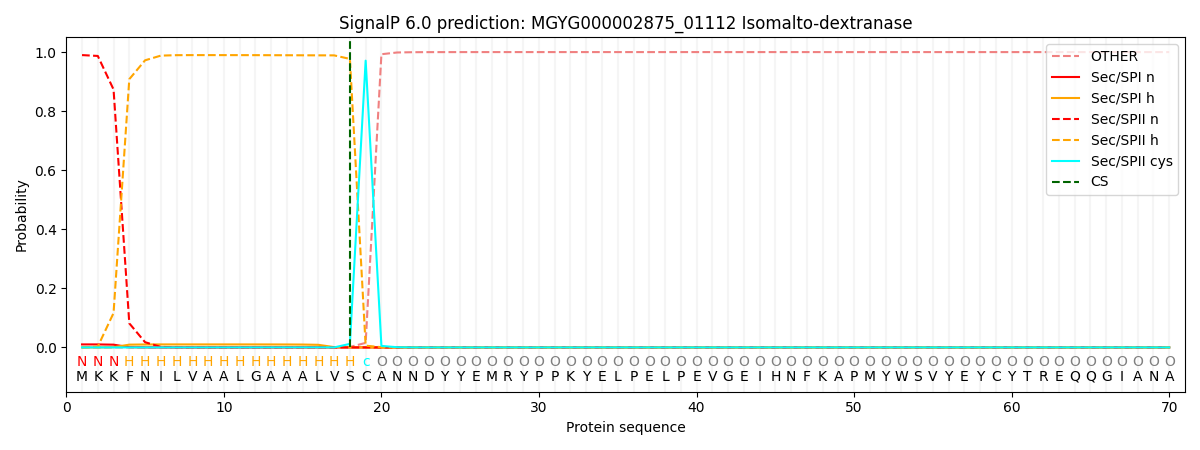
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000011 | 0.009979 | 0.990054 | 0.000003 | 0.000004 | 0.000003 |
