You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002885_01610
You are here: Home > Sequence: MGYG000002885_01610
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
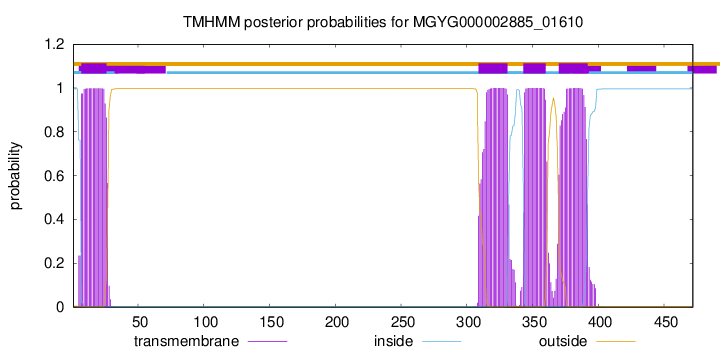
TMHMM annotations
Basic Information help
| Species | Pseudomonas_E juntendi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_E; Pseudomonas_E juntendi | |||||||||||
| CAZyme ID | MGYG000002885_01610 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54; End: 1472 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5309 | Scw11 | 9.19e-106 | 1 | 296 | 1 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| cd06435 | CESA_NdvC_like | 8.52e-26 | 424 | 472 | 1 | 49 | NdvC_like proteins in this family are putative bacterial beta-(1,6)-glucosyltransferase. NdvC_like proteins in this family are putative bacterial beta-(1,6)-glucosyltransferase. Bradyrhizobium japonicum synthesizes periplasmic cyclic beta-(1,3),beta-(1,6)-D-glucans during growth under hypoosmotic conditions. Two genes (ndvB, ndvC) are involved in the beta-(1, 3), beta-(1,6)-glucan synthesis. The ndvC mutant strain resulted in synthesis of altered cyclic beta-glucans composed almost entirely of beta-(1, 3)-glycosyl linkages. The periplasmic cyclic beta-(1,3),beta-(1,6)-D-glucans function for osmoregulation. The ndvC mutation also affects the ability of the bacteria to establish a successful symbiotic interaction with host plant. Thus, the beta-glucans may function as suppressors of a host defense response. |
| COG1215 | BcsA | 1.90e-11 | 373 | 472 | 10 | 104 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06423 | CESA_like | 7.19e-10 | 425 | 463 | 1 | 38 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| PRK11204 | PRK11204 | 1.04e-08 | 403 | 463 | 38 | 95 | N-glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOH68702.1 | 0.0 | 1 | 472 | 1 | 472 |
| QKL00875.1 | 0.0 | 1 | 472 | 1 | 472 |
| QDR67129.1 | 0.0 | 1 | 472 | 1 | 472 |
| QEQ86882.1 | 0.0 | 1 | 472 | 1 | 472 |
| APE97604.1 | 0.0 | 1 | 472 | 1 | 472 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NTP7 | 1.73e-19 | 26 | 299 | 371 | 671 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
| Q2U492 | 1.73e-19 | 26 | 299 | 371 | 671 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
| D4B2W4 | 5.96e-18 | 48 | 301 | 26 | 298 | Glucan 1,3-beta-glucosidase ARB_02797 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02797 PE=1 SV=1 |
| Q5B430 | 2.85e-17 | 42 | 305 | 347 | 641 | Glucan endo-1,3-beta-glucosidase btgC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=btgC PE=2 SV=1 |
| P53334 | 3.42e-17 | 40 | 295 | 133 | 386 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
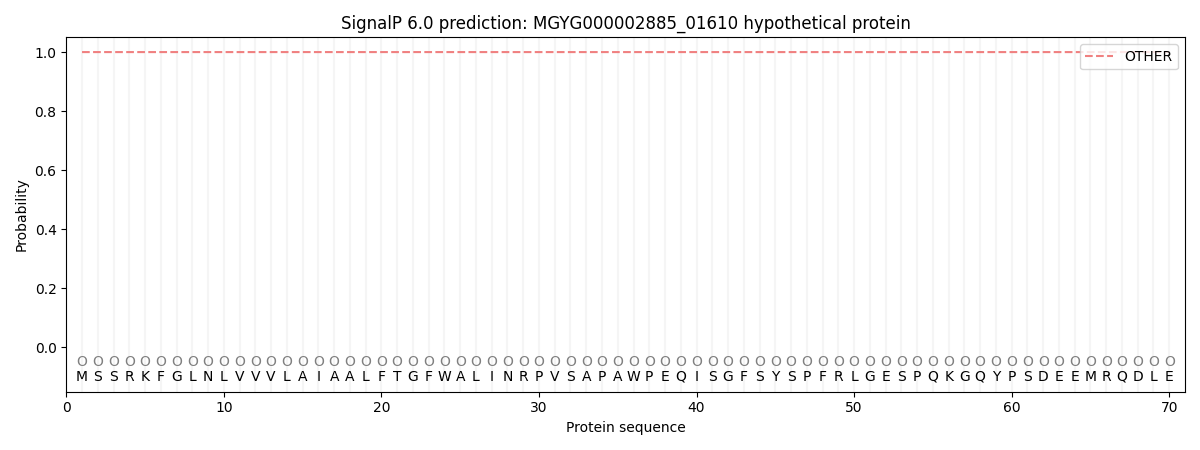
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000041 | 0.000003 | 0.000001 | 0.000000 | 0.000000 | 0.000001 |