You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002886_01693
You are here: Home > Sequence: MGYG000002886_01693
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
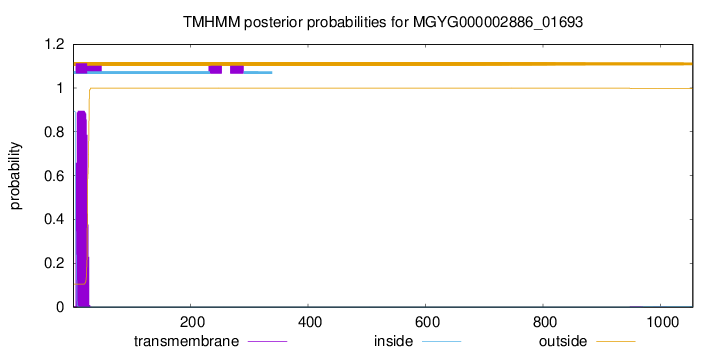
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia | |||||||||||
| CAZyme ID | MGYG000002886_01693 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13093; End: 16260 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 204 | 303 | 1.6e-17 | 0.8064516129032258 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.33e-13 | 208 | 304 | 18 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 5.58e-06 | 208 | 291 | 29 | 126 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 3.21e-05 | 208 | 304 | 28 | 133 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| COG3408 | GDB1 | 0.002 | 459 | 714 | 105 | 374 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| COG3459 | COG3459 | 0.002 | 517 | 710 | 496 | 700 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJP19414.1 | 0.0 | 1 | 1055 | 1 | 1055 |
| QNG72305.1 | 0.0 | 1 | 1055 | 1 | 1055 |
| QNG96179.1 | 0.0 | 1 | 1055 | 1 | 1055 |
| QNG68187.1 | 0.0 | 1 | 1055 | 1 | 1055 |
| QJC74097.1 | 0.0 | 1 | 1055 | 1 | 1055 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
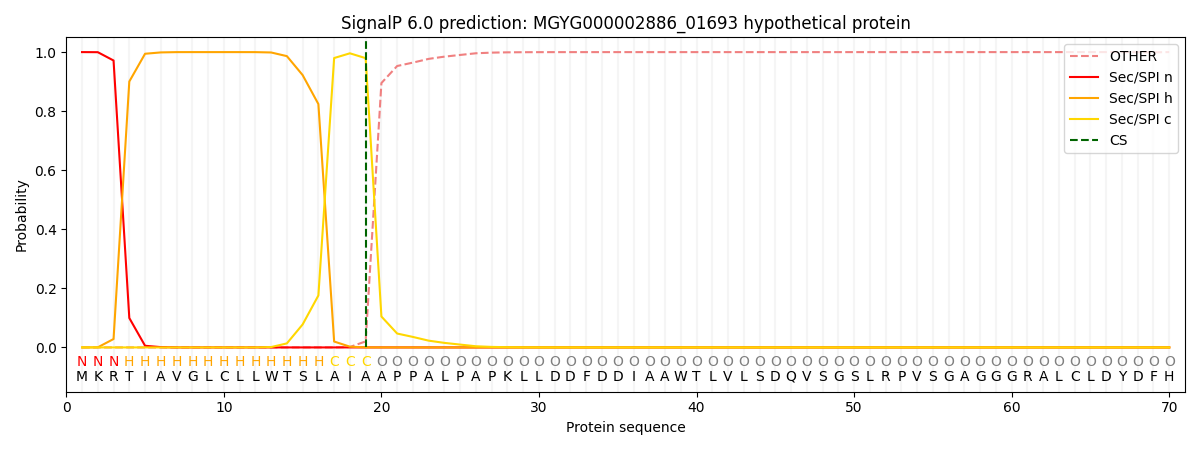
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000397 | 0.998759 | 0.000216 | 0.000214 | 0.000197 | 0.000187 |