You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002886_03042
You are here: Home > Sequence: MGYG000002886_03042
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia | |||||||||||
| CAZyme ID | MGYG000002886_03042 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Alpha-N-acetylgalactosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2426; End: 3781 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 29 | 440 | 1.5e-170 | 0.9874686716791979 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.76e-19 | 28 | 424 | 1 | 335 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 3.56e-10 | 31 | 133 | 1 | 92 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| smart00859 | Semialdhyde_dh | 0.002 | 32 | 130 | 1 | 95 | Semialdehyde dehydrogenase, NAD binding domain. The semialdehyde dehydrogenase family is found in N-acetyl-glutamine semialdehyde dehydrogenase (AgrC), which is involved in arginine biosynthesis, and aspartate-semialdehyde dehydrogenase, an enzyme involved in the biosynthesis of various amino acids from aspartate. This family is also found in yeast and fungal Arg5,6 protein, which is cleaved into the enzymes N-acety-gamma-glutamyl-phosphate reductase and acetylglutamate kinase. These are also involved in arginine biosynthesis. All proteins in this entry contain a NAD binding region of semialdehyde dehydrogenase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYZ72572.1 | 0.0 | 1 | 451 | 1 | 451 |
| QJP21687.1 | 0.0 | 1 | 451 | 1 | 451 |
| QJC72949.1 | 0.0 | 1 | 451 | 1 | 451 |
| QHE22867.1 | 0.0 | 1 | 451 | 1 | 451 |
| QBL46882.1 | 0.0 | 1 | 451 | 1 | 451 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2IXA_A | 5.44e-157 | 29 | 451 | 19 | 444 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 6T2B_A | 2.30e-75 | 30 | 444 | 42 | 444 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 3E18_A | 7.62e-09 | 29 | 203 | 4 | 171 | CRYSTALSTRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua],3E18_B CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua] |
| 6A3F_A | 1.59e-08 | 82 | 237 | 72 | 207 | Levoglucosandehydrogenase, apo form [Pseudarthrobacter phenanthrenivorans Sphe3],6A3F_B Levoglucosan dehydrogenase, apo form [Pseudarthrobacter phenanthrenivorans Sphe3],6A3G_A Levoglucosan dehydrogenase, complex with NADH [Pseudarthrobacter phenanthrenivorans Sphe3],6A3G_B Levoglucosan dehydrogenase, complex with NADH [Pseudarthrobacter phenanthrenivorans Sphe3],6A3G_C Levoglucosan dehydrogenase, complex with NADH [Pseudarthrobacter phenanthrenivorans Sphe3],6A3G_D Levoglucosan dehydrogenase, complex with NADH [Pseudarthrobacter phenanthrenivorans Sphe3],6A3I_A Levoglucosan dehydrogenase, complex with NADH and levoglucosan [Pseudarthrobacter phenanthrenivorans Sphe3],6A3I_B Levoglucosan dehydrogenase, complex with NADH and levoglucosan [Pseudarthrobacter phenanthrenivorans Sphe3],6A3I_C Levoglucosan dehydrogenase, complex with NADH and levoglucosan [Pseudarthrobacter phenanthrenivorans Sphe3],6A3I_D Levoglucosan dehydrogenase, complex with NADH and levoglucosan [Pseudarthrobacter phenanthrenivorans Sphe3],6A3J_A Levoglucosan dehydrogenase, complex with NADH and L-sorbose [Pseudarthrobacter phenanthrenivorans Sphe3],6A3J_B Levoglucosan dehydrogenase, complex with NADH and L-sorbose [Pseudarthrobacter phenanthrenivorans Sphe3],6A3J_C Levoglucosan dehydrogenase, complex with NADH and L-sorbose [Pseudarthrobacter phenanthrenivorans Sphe3],6A3J_D Levoglucosan dehydrogenase, complex with NADH and L-sorbose [Pseudarthrobacter phenanthrenivorans Sphe3] |
| 3EC7_A | 4.10e-07 | 27 | 154 | 20 | 138 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2FLK4 | 0.0 | 1 | 451 | 4 | 454 | Glycosyl hydrolase family 109 protein OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=Smlt4431 PE=3 SV=1 |
| A4Q8F7 | 2.98e-156 | 29 | 451 | 19 | 444 | Alpha-N-acetylgalactosaminidase OS=Elizabethkingia meningoseptica OX=238 GN=nagA PE=1 SV=1 |
| A4Q8G1 | 2.12e-144 | 23 | 446 | 45 | 467 | Alpha-N-acetylgalactosaminidase OS=Tannerella forsythia OX=28112 GN=nagA PE=3 SV=1 |
| A6LB54 | 2.59e-142 | 30 | 446 | 49 | 465 | Glycosyl hydrolase family 109 protein OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=BDI_1155 PE=3 SV=1 |
| A8H2K3 | 2.76e-94 | 31 | 444 | 53 | 451 | Glycosyl hydrolase family 109 protein OS=Shewanella pealeana (strain ATCC 700345 / ANG-SQ1) OX=398579 GN=Spea_1465 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
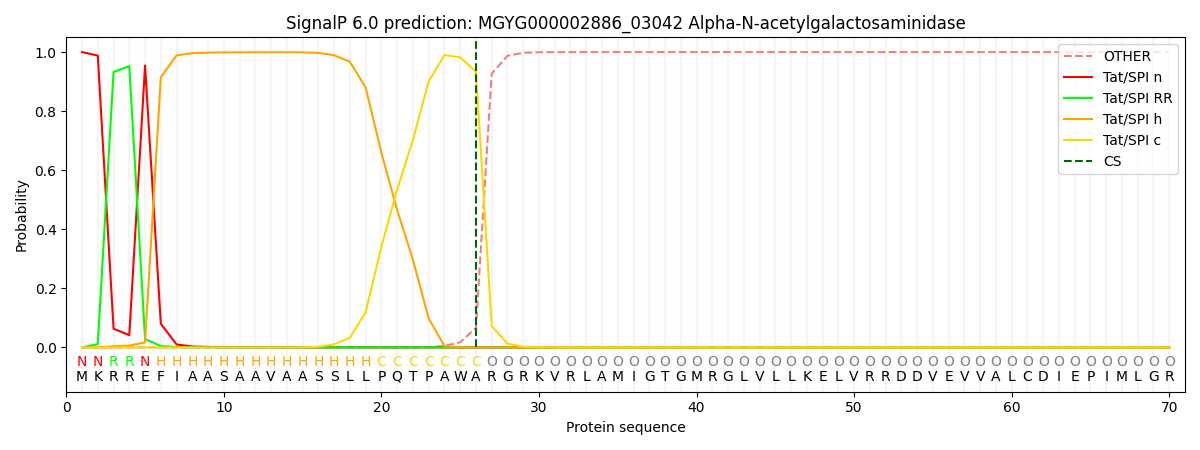
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 1.000004 | 0.000000 | 0.000000 |
