You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002889_02320
You are here: Home > Sequence: MGYG000002889_02320
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pantoea sp002920175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pantoea; Pantoea sp002920175 | |||||||||||
| CAZyme ID | MGYG000002889_02320 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54946; End: 55419 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 33 | 153 | 1.3e-23 | 0.8518518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13400 | LT_IagB-like | 1.55e-49 | 37 | 151 | 1 | 107 | Escherichia coli invasion protein IagB and similar proteins. Lytic transglycosylase-like protein, similar to Escherichia coli invasion protein IagB. IagB is encoded within a pathogenicity island in Salmonella enterica and has been shown to degrade polymeric peptidoglycan. IagB-like invasion proteins are implicated in the invasion of eukaryotic host cells by bacteria. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Members of this family resemble the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| PRK15328 | PRK15328 | 1.65e-39 | 27 | 156 | 19 | 144 | type III secretion system invasion protein IagB. |
| pfam01464 | SLT | 5.27e-22 | 30 | 146 | 1 | 110 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| PRK13722 | PRK13722 | 3.87e-17 | 26 | 151 | 19 | 142 | lytic transglycosylase; Provisional |
| cd00254 | LT-like | 6.27e-13 | 42 | 151 | 2 | 110 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBJ48260.1 | 9.41e-110 | 1 | 157 | 1 | 157 |
| QZY97764.1 | 8.98e-99 | 1 | 157 | 1 | 157 |
| QIA54850.1 | 7.38e-98 | 1 | 157 | 1 | 157 |
| AVF38279.1 | 5.05e-83 | 12 | 156 | 4 | 148 |
| QTP13580.1 | 3.99e-78 | 1 | 156 | 1 | 158 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XP8_A | 5.35e-10 | 29 | 115 | 3 | 86 | Structureof EtgA D60N mutant [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8X6H3 | 1.21e-27 | 29 | 154 | 28 | 156 | Peptidoglycan-binding-like protein OS=Escherichia coli O157:H7 OX=83334 GN=pbl PE=3 SV=1 |
| E1WAC2 | 1.13e-26 | 27 | 156 | 19 | 144 | Invasion protein IagB OS=Salmonella typhimurium (strain SL1344) OX=216597 GN=iagB PE=3 SV=1 |
| P0CL15 | 1.13e-26 | 27 | 156 | 19 | 144 | Invasion protein IagB OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=iagB PE=3 SV=1 |
| P43018 | 9.02e-26 | 27 | 154 | 19 | 142 | Invasion protein IagB OS=Salmonella typhi OX=90370 GN=iagB PE=3 SV=1 |
| Q07568 | 2.91e-25 | 27 | 156 | 18 | 143 | Protein IpgF OS=Shigella flexneri OX=623 GN=ipgF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
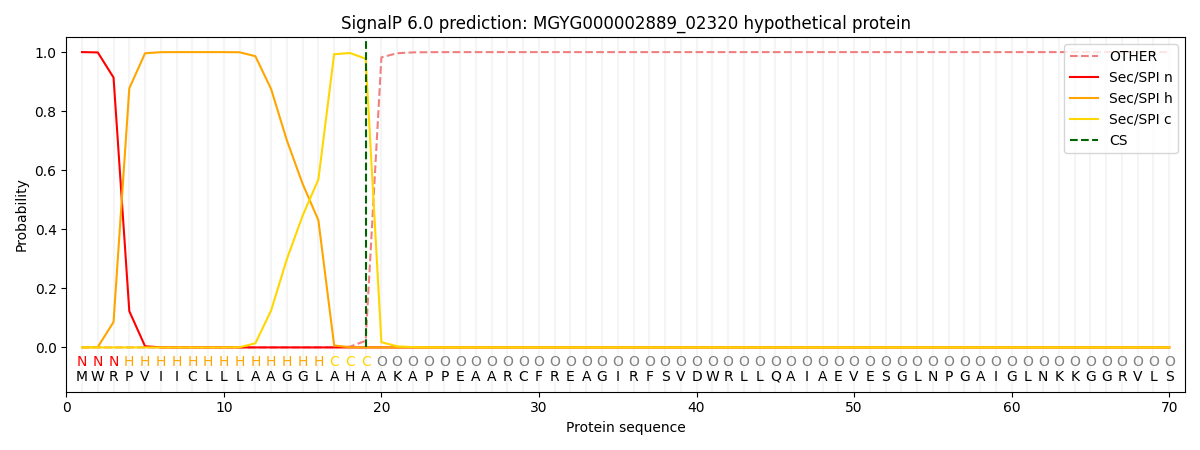
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000242 | 0.999129 | 0.000166 | 0.000157 | 0.000146 | 0.000137 |
