You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002889_04048
You are here: Home > Sequence: MGYG000002889_04048
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
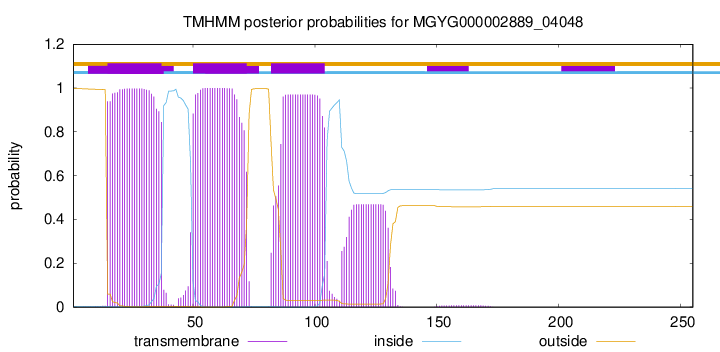
TMHMM annotations
Basic Information help
| Species | Pantoea sp002920175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pantoea; Pantoea sp002920175 | |||||||||||
| CAZyme ID | MGYG000002889_04048 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14; End: 781 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 2 | 227 | 3.1e-41 | 0.43333333333333335 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13279 | arnT | 1.63e-107 | 1 | 252 | 300 | 551 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1807 | ArnT | 2.43e-14 | 1 | 215 | 303 | 523 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDY43895.1 | 9.59e-131 | 2 | 255 | 300 | 553 |
| QFS62523.1 | 2.07e-129 | 3 | 255 | 301 | 551 |
| QZY97503.1 | 8.31e-129 | 3 | 255 | 301 | 551 |
| QZY92886.1 | 1.18e-128 | 3 | 255 | 301 | 551 |
| QCP62241.1 | 6.24e-121 | 2 | 255 | 300 | 553 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2VBI7 | 1.70e-83 | 3 | 255 | 301 | 553 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| A6TF96 | 2.28e-83 | 3 | 251 | 301 | 548 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=arnT PE=3 SV=1 |
| B5XTL1 | 1.79e-82 | 3 | 251 | 301 | 548 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Klebsiella pneumoniae (strain 342) OX=507522 GN=arnT PE=3 SV=1 |
| A4WAM1 | 1.09e-77 | 3 | 255 | 301 | 553 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Enterobacter sp. (strain 638) OX=399742 GN=arnT PE=3 SV=1 |
| Q2NRV9 | 2.85e-77 | 3 | 251 | 301 | 545 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Sodalis glossinidius (strain morsitans) OX=343509 GN=arnT2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
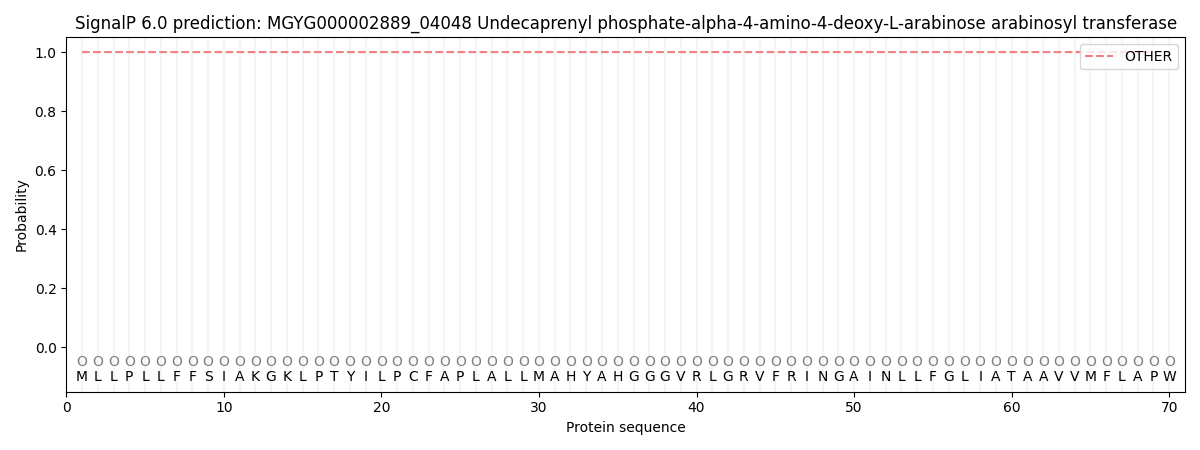
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000019 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |