You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002907_00870
You are here: Home > Sequence: MGYG000002907_00870
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerosporomusa sp900542835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Sporomusales; Acetonemaceae; Anaerosporomusa; Anaerosporomusa sp900542835 | |||||||||||
| CAZyme ID | MGYG000002907_00870 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47759; End: 49300 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 306 | 454 | 2.4e-30 | 0.41254125412541254 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 2.95e-18 | 323 | 474 | 181 | 325 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 3.14e-13 | 299 | 445 | 129 | 257 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBB91187.1 | 3.66e-179 | 17 | 510 | 34 | 531 |
| AKL94176.1 | 8.83e-176 | 32 | 511 | 41 | 526 |
| QUH24907.1 | 3.09e-173 | 7 | 511 | 10 | 520 |
| AOY76751.1 | 5.99e-173 | 13 | 511 | 21 | 523 |
| ARE87201.1 | 5.99e-173 | 13 | 511 | 21 | 523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVD_A | 1.55e-29 | 300 | 511 | 78 | 277 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 1.55e-29 | 300 | 511 | 78 | 277 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 1.80e-29 | 300 | 511 | 78 | 277 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2VI0_A | 3.93e-29 | 300 | 490 | 78 | 273 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 3.93e-29 | 300 | 511 | 78 | 277 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 7.50e-27 | 300 | 511 | 100 | 299 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| O05512 | 1.70e-09 | 315 | 452 | 177 | 301 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P16699 | 5.78e-08 | 316 | 408 | 180 | 271 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| G2Q4H7 | 1.58e-06 | 317 | 424 | 318 | 411 | Mannan endo-1,4-beta-mannosidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Man26A PE=1 SV=1 |
| P49424 | 5.78e-06 | 301 | 407 | 180 | 285 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
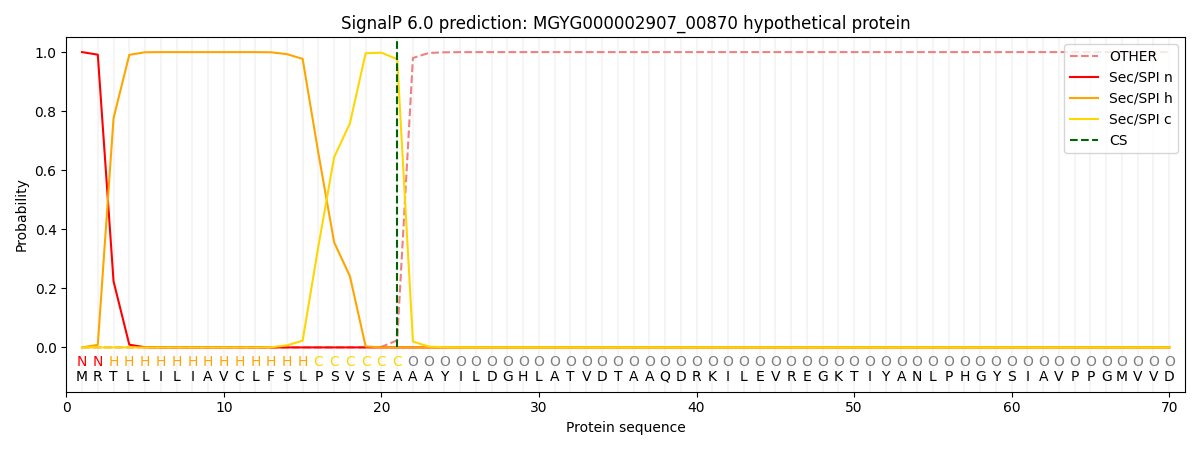
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000245 | 0.999070 | 0.000193 | 0.000168 | 0.000163 | 0.000148 |
