You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002922_01301
You are here: Home > Sequence: MGYG000002922_01301
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900541925 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900541925 | |||||||||||
| CAZyme ID | MGYG000002922_01301 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64555; End: 66861 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 65 | 377 | 3.8e-53 | 0.9739776951672863 |
| CE15 | 418 | 764 | 1.5e-24 | 0.9405204460966543 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12146 | Hydrolase_4 | 4.56e-11 | 534 | 664 | 6 | 119 | Serine aminopeptidase, S33. This domain is found in bacteria and eukaryotes and is approximately 110 amino acids in length. It is found in association with pfam00561. The majority of the members in this family carry the exopeptidase active-site residues of Ser-122, Asp-239 and His-269 as in UniProtKB:Q7ZWC2. |
| COG1506 | DAP2 | 2.80e-10 | 489 | 649 | 348 | 503 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam00561 | Abhydrolase_1 | 9.77e-09 | 533 | 694 | 1 | 131 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| COG0596 | MhpC | 1.65e-07 | 527 | 761 | 16 | 220 | Pimeloyl-ACP methyl ester carboxylesterase [Coenzyme transport and metabolism, General function prediction only]. |
| COG0412 | DLH | 1.53e-06 | 507 | 648 | 3 | 141 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKJ30743.1 | 5.41e-305 | 1 | 766 | 8 | 779 |
| BBD44973.1 | 7.25e-148 | 25 | 766 | 50 | 794 |
| SCM59684.1 | 1.55e-144 | 42 | 767 | 22 | 740 |
| QOY89374.1 | 1.20e-138 | 45 | 765 | 51 | 720 |
| AEL25656.1 | 9.12e-127 | 45 | 765 | 62 | 760 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 1.25e-29 | 65 | 379 | 24 | 365 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
| 6EHN_A | 5.22e-26 | 65 | 352 | 14 | 354 | Structuralinsight into a promiscuous CE15 esterase from the marine bacterial metagenome [unidentified prokaryotic organism] |
| 6GRW_A | 2.43e-25 | 65 | 350 | 7 | 350 | GlucuronoylEsterase from Opitutus terrae (Au derivative) [Opitutus terrae PB90-1],6GS0_A Native Glucuronoyl Esterase from Opitutus terrae [Opitutus terrae PB90-1] |
| 6SZ0_A | 3.07e-25 | 65 | 350 | 25 | 368 | Theglucuronoyl esterase OtCE15A H408A variant from Opitutus terrae [Opitutus terrae PB90-1],6SZ4_A The glucuronoyl esterase OtCE15A H408A variant from Opitutus terrae in complex with, and covalently linked to, D-glucuronate [Opitutus terrae PB90-1] |
| 6SYU_A | 3.07e-25 | 65 | 350 | 25 | 368 | Thewild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with xylobiose [Opitutus terrae],6T0I_A The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with the aldotetrauronic acid XUX [Opitutus terrae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A0K2VM55 | 4.42e-25 | 65 | 352 | 42 | 382 | Carbohydrate esterase MZ0003 OS=Unknown prokaryotic organism OX=2725 GN=MZ0003 PE=1 SV=1 |
| A0A0A7EQR3 | 4.04e-24 | 64 | 320 | 111 | 375 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
| D8QLP9 | 3.01e-23 | 61 | 362 | 32 | 336 | 4-O-methyl-glucuronoyl methylesterase OS=Schizophyllum commune (strain H4-8 / FGSC 9210) OX=578458 GN=SCHCODRAFT_238770 PE=1 SV=1 |
| K5XDZ6 | 4.78e-23 | 61 | 360 | 40 | 346 | 4-O-methyl-glucuronoyl methylesterase OS=Phanerochaete carnosa (strain HHB-10118-sp) OX=650164 GN=PHACADRAFT_247750 PE=1 SV=1 |
| B2ABS0 | 7.81e-23 | 50 | 323 | 106 | 391 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
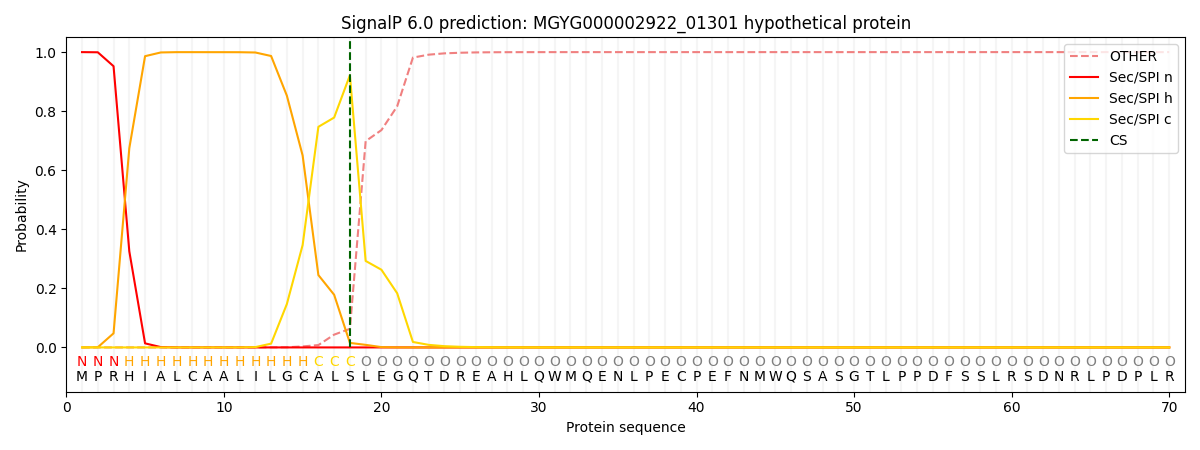
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000334 | 0.998606 | 0.000534 | 0.000169 | 0.000165 | 0.000157 |
