You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002933_01986
Basic Information
help
| Species |
Phocaeicola sp900540105
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900540105
|
| CAZyme ID |
MGYG000002933_01986
|
| CAZy Family |
CBM32 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002933 |
3120707 |
MAG |
Estonia |
Europe |
|
| Gene Location |
Start: 13939;
End: 14817
Strand: -
|
No EC number prediction in MGYG000002933_01986.
| Family |
Start |
End |
Evalue |
family coverage |
| CBM32 |
146 |
277 |
8.4e-17 |
0.9193548387096774 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00754
|
F5_F8_type_C |
1.32e-12 |
144 |
276 |
2 |
127 |
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057
|
FA58C |
9.34e-06 |
157 |
234 |
26 |
97 |
Substituted updates: Jan 31, 2002 |
This protein is predicted as SP
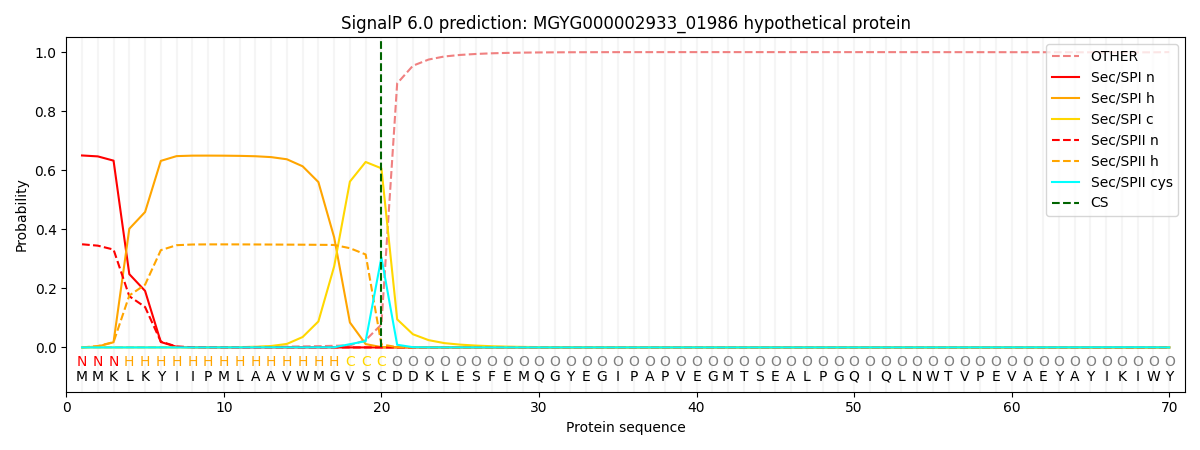
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.001852
|
0.644141
|
0.353342
|
0.000246
|
0.000214
|
0.000204
|
There is no transmembrane helices in MGYG000002933_01986.

