You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002933_02268
You are here: Home > Sequence: MGYG000002933_02268
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900540105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900540105 | |||||||||||
| CAZyme ID | MGYG000002933_02268 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12232; End: 13644 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 35 | 362 | 4.6e-58 | 0.8526011560693642 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3669 | AfuC | 6.82e-35 | 30 | 465 | 8 | 430 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| pfam01120 | Alpha_L_fucos | 6.42e-23 | 77 | 362 | 79 | 326 | Alpha-L-fucosidase. |
| smart00812 | Alpha_L_fucos | 3.99e-20 | 77 | 361 | 76 | 327 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| cd07436 | PHP_PolX | 0.001 | 78 | 108 | 171 | 202 | Polymerase and Histidinol Phosphatase domain of bacterial polymerase X. The bacterial/archaeal X-family DNA polymerases (PolXs) have a PHP domain at their C-terminus. The bacterial/archaeal PolX core domain and PHP domain interact with each other and together are involved in metal dependent 3'-5' exonuclease activity. The PHP (also called histidinol phosphatase-2/HIS2) domain is associated with several types of DNA polymerases, such as PolIIIA and family X DNA polymerases, stand alone histidinol phosphate phosphatases (HisPPases), and a number of uncharacterized protein families. The PHP domain has four conserved sequence motifs and contains an invariant histidine that is involved in metal ion coordination. PolX is found in all kingdoms, however bacterial PolXs have a completely different domain structure from eukaryotic PolXs. Bacterial PolX has an extended conformation in contrast to the common closed 'right hand' conformation for DNA polymerases. This extended conformation is stabilized by the PHP domain. The PHP domain of PolX is structurally homologous to other members of the PHP family that has a distorted (beta/alpha)7 barrel fold with a trinuclear metal site on the C-terminal side of the barrel. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO23619.1 | 2.43e-303 | 13 | 467 | 16 | 469 |
| QWO87069.1 | 1.34e-270 | 33 | 470 | 30 | 466 |
| QWO94329.1 | 1.34e-270 | 33 | 470 | 30 | 466 |
| QWO96892.1 | 1.34e-270 | 33 | 470 | 30 | 466 |
| QWP71663.1 | 1.34e-270 | 33 | 470 | 30 | 466 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GZA_A | 1.06e-252 | 27 | 467 | 4 | 442 | Crystalstructure of putative alpha-L-fucosidase (NP_812709.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.60 A resolution [Bacteroides thetaiotaomicron VPI-5482],3GZA_B Crystal structure of putative alpha-L-fucosidase (NP_812709.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.60 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 5K9H_A | 2.59e-68 | 30 | 465 | 36 | 463 | Crystalstructure of a glycoside hydrolase 29 family member from an unknown rumen bacterium [unidentified] |
| 4ZRX_A | 5.25e-64 | 33 | 466 | 32 | 459 | Crystalstructure of a putative alpha-L-fucosidase (BACOVA_04357) from Bacteroides ovatus ATCC 8483 at 1.59 A resolution [Bacteroides ovatus ATCC 8483] |
| 3EYP_A | 6.65e-62 | 30 | 465 | 4 | 454 | Crystalstructure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],3EYP_B Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],4OUE_A Crystal structure of an a-L-Fucosidase GH29 from Bacteroides thetaiotaomicron (BT2192) in complex with IPTG [Bacteroides thetaiotaomicron VPI-5482],4OUE_B Crystal structure of an a-L-Fucosidase GH29 from Bacteroides thetaiotaomicron (BT2192) in complex with IPTG [Bacteroides thetaiotaomicron VPI-5482],4OZO_A Crystal structure of an a-L-fucosidase GH29 from Bacteroides thetaiotaomicron (BT2192) in complex with oNPTG [Bacteroides thetaiotaomicron VPI-5482],4OZO_B Crystal structure of an a-L-fucosidase GH29 from Bacteroides thetaiotaomicron (BT2192) in complex with oNPTG [Bacteroides thetaiotaomicron VPI-5482] |
| 3UES_A | 1.60e-49 | 33 | 459 | 19 | 470 | Crystalstructure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088],3UES_B Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7XUR3 | 7.04e-63 | 31 | 465 | 38 | 476 | Putative alpha-L-fucosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0560400 PE=3 SV=2 |
| Q8GW72 | 3.08e-61 | 31 | 465 | 36 | 477 | Alpha-L-fucosidase 1 OS=Arabidopsis thaliana OX=3702 GN=FUC1 PE=1 SV=2 |
| Q99KR8 | 4.29e-07 | 65 | 361 | 85 | 351 | Plasma alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca2 PE=1 SV=1 |
| Q6AYS4 | 2.30e-06 | 72 | 361 | 93 | 349 | Plasma alpha-L-fucosidase OS=Rattus norvegicus OX=10116 GN=Fuca2 PE=2 SV=1 |
| P10901 | 4.05e-06 | 39 | 206 | 81 | 223 | Alpha-L-fucosidase OS=Dictyostelium discoideum OX=44689 GN=alfA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
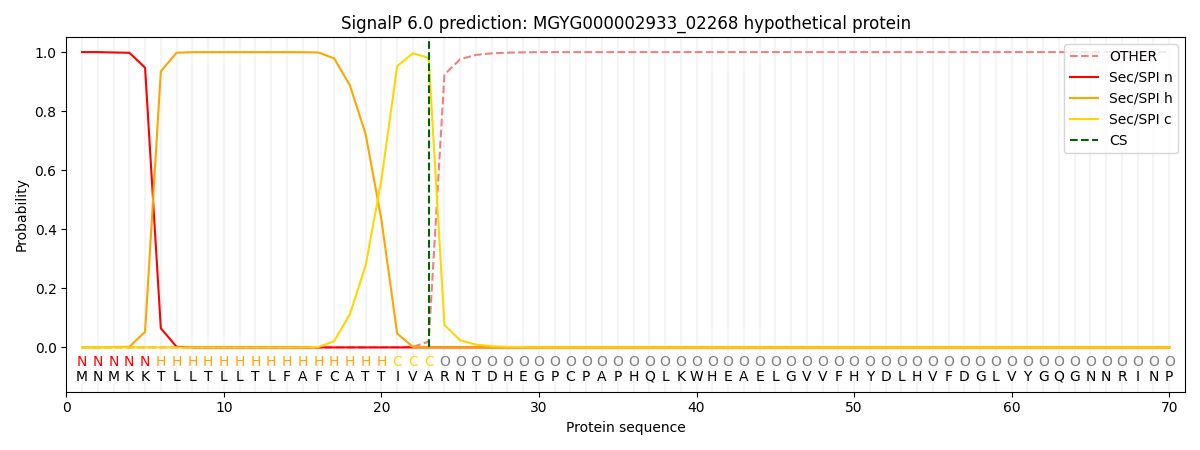
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000206 | 0.999168 | 0.000193 | 0.000148 | 0.000143 | 0.000131 |
