You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002933_02429
You are here: Home > Sequence: MGYG000002933_02429
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900540105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900540105 | |||||||||||
| CAZyme ID | MGYG000002933_02429 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase ArbA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12860; End: 13828 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 23 | 306 | 4.5e-113 | 0.9966555183946488 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18616 | GH43_ABN-like | 8.58e-156 | 25 | 301 | 1 | 291 | Glycosyl hydrolase family 43 such as arabinan endo-1 5-alpha-L-arabinosidase. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activity. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08998 | GH43_Arb43a-like | 1.90e-85 | 34 | 302 | 3 | 278 | Glycosyl hydrolase family 43 protein such as Bacillus subtilis subsp. subtilis str. 168 endo-alpha-1,5-L-arabinanase Arb43A. This glycosyl hydrolase family 43 (GH43) subgroup belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. Many of these enzymes such as the Bacillus subtilis arabinanase Abn2, that hydrolyzes sugar beet arabinan (branched), linear alpha-1,5-L-arabinan and pectin, are different from other arabinases; they are organized into two different domains with a divalent metal cluster close to the catalytic residues to guarantee the correct protonation state of the catalytic residues and consequently the enzyme activity. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam04616 | Glyco_hydro_43 | 3.50e-83 | 23 | 306 | 1 | 281 | Glycosyl hydrolases family 43. The glycosyl hydrolase family 43 contains members that are arabinanases. Arabinanases hydrolyze the alpha-1,5-linked L-arabinofuranoside backbone of plant cell wall arabinans. The structure of arabinanase Arb43A from Cellvibrio japonicus reveals a five-bladed beta-propeller fold. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08988 | GH43_ABN | 9.36e-73 | 33 | 301 | 1 | 277 | Glycosyl hydrolase family 43. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with alpha-L-arabinofuranosidase (ABF; EC 3.2.1.55) and endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08999 | GH43_ABN-like | 1.02e-59 | 25 | 307 | 1 | 284 | Glycosyl hydrolase family 43 protein such as endo-alpha-L-arabinanase. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with alpha-L-arabinofuranosidase (ABF; EC 3.2.1.55) and endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT89097.1 | 1.62e-197 | 1 | 322 | 1 | 325 |
| ALJ59878.1 | 6.58e-197 | 1 | 322 | 1 | 325 |
| QDO68151.1 | 2.20e-195 | 1 | 322 | 1 | 325 |
| QDO68166.1 | 3.77e-189 | 20 | 322 | 32 | 334 |
| QBJ18534.1 | 7.60e-189 | 20 | 320 | 32 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GYD_B | 5.76e-40 | 34 | 308 | 6 | 301 | Structureof Cellvibrio cellulosa alpha-L-arabinanase [Cellvibrio japonicus] |
| 1UV4_A | 5.10e-39 | 32 | 307 | 12 | 292 | NativeBacillus subtilis Arabinanase Arb43A [Bacillus subtilis] |
| 1GYE_B | 8.53e-39 | 34 | 308 | 7 | 302 | Structureof Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose [Cellvibrio japonicus] |
| 1GYH_A | 9.13e-39 | 34 | 308 | 9 | 304 | Structureof D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus],1GYH_B Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus],1GYH_C Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus],1GYH_D Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus],1GYH_E Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus],1GYH_F Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant [Cellvibrio japonicus] |
| 6B7K_A | 1.79e-37 | 29 | 307 | 18 | 300 | GH43Endo-Arabinanase from Bacillus licheniformis [Bacillus licheniformis DSM 13 = ATCC 14580],6B7K_B GH43 Endo-Arabinanase from Bacillus licheniformis [Bacillus licheniformis DSM 13 = ATCC 14580],6B7K_C GH43 Endo-Arabinanase from Bacillus licheniformis [Bacillus licheniformis DSM 13 = ATCC 14580],6B7K_D GH43 Endo-Arabinanase from Bacillus licheniformis [Bacillus licheniformis DSM 13 = ATCC 14580] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P95470 | 1.26e-38 | 34 | 308 | 38 | 333 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase ArbA OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=arbA PE=1 SV=1 |
| P94522 | 5.60e-38 | 32 | 307 | 42 | 322 | Extracellular endo-alpha-(1->5)-L-arabinanase 1 OS=Bacillus subtilis (strain 168) OX=224308 GN=abnA PE=1 SV=3 |
| B3EYM8 | 1.35e-36 | 34 | 306 | 27 | 311 | Intracellular endo-alpha-(1->5)-L-arabinanase OS=Geobacillus stearothermophilus OX=1422 GN=abnB PE=1 SV=1 |
| Q93HT9 | 4.93e-36 | 34 | 306 | 27 | 311 | Intracellular endo-alpha-(1->5)-L-arabinanase OS=Geobacillus thermodenitrificans OX=33940 GN=abn-ts PE=1 SV=1 |
| B8NDL1 | 3.44e-30 | 31 | 308 | 31 | 319 | Probable arabinan endo-1,5-alpha-L-arabinosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abnA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
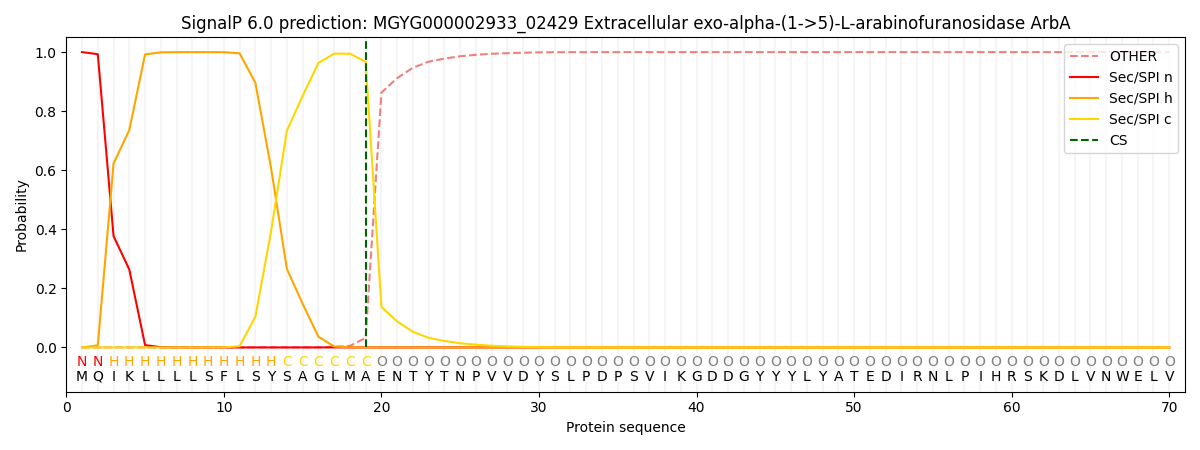
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000541 | 0.998748 | 0.000185 | 0.000183 | 0.000162 | 0.000149 |
