You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002933_02497
You are here: Home > Sequence: MGYG000002933_02497
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900540105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900540105 | |||||||||||
| CAZyme ID | MGYG000002933_02497 | |||||||||||
| CAZy Family | PL42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 261; End: 1547 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL42 | 34 | 326 | 4e-127 | 0.9966216216216216 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15892 | BNR_4 | 8.95e-126 | 51 | 313 | 1 | 272 | BNR repeat-containing family member. BNR_4 is a family which carries the unique sequence motif SxDxGxTW which is so characteristic of the repeats of the BNR family, pfam02012. It is unclear whether or not this unit is repeated throughout the sequences of this family, but if it is then the family is likely to be bacterial neuraminidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY36173.1 | 5.50e-242 | 1 | 424 | 2 | 424 |
| QJR78280.1 | 1.55e-235 | 1 | 424 | 1 | 423 |
| QUT85916.1 | 4.04e-235 | 16 | 424 | 3 | 411 |
| AII65648.1 | 4.04e-235 | 16 | 424 | 3 | 411 |
| QJR61378.1 | 6.29e-235 | 1 | 424 | 1 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MUK_A | 9.40e-233 | 18 | 424 | 18 | 424 | GlycosideHydrolase BT3686 [Bacteroides thetaiotaomicron VPI-5482],5MUL_A Glycoside Hydrolase BT3686 bound to Glucuronic Acid [Bacteroides thetaiotaomicron VPI-5482] |
| 5MUM_A | 1.25e-228 | 21 | 424 | 26 | 429 | GlycosideHydrolase BACINT_00347 [Bacteroides intestinalis DSM 17393] |
| 5MVH_A | 2.83e-227 | 18 | 426 | 18 | 426 | GlycosideHydrolase BACCELL_00856 [Bacteroides cellulosilyticus DSM 14838] |
| 4IRT_A | 3.06e-227 | 20 | 424 | 3 | 407 | Crystalstructure of a putative neuraminidase (BACOVA_03493) from Bacteroides ovatus ATCC 8483 at 1.74 A resolution [Bacteroides ovatus ATCC 8483] |
| 7ESK_A | 2.92e-37 | 35 | 298 | 23 | 304 | ChainA, L-Rhamnose-alpha-1,4-D-glucuronate lyase [Fusarium oxysporum],7ESM_A Chain A, L-rhamnose-alpha-1,4-D-glucuronate lyase [Fusarium oxysporum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WF05 | 3.55e-06 | 38 | 307 | 67 | 375 | Ulvan lyase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_29 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
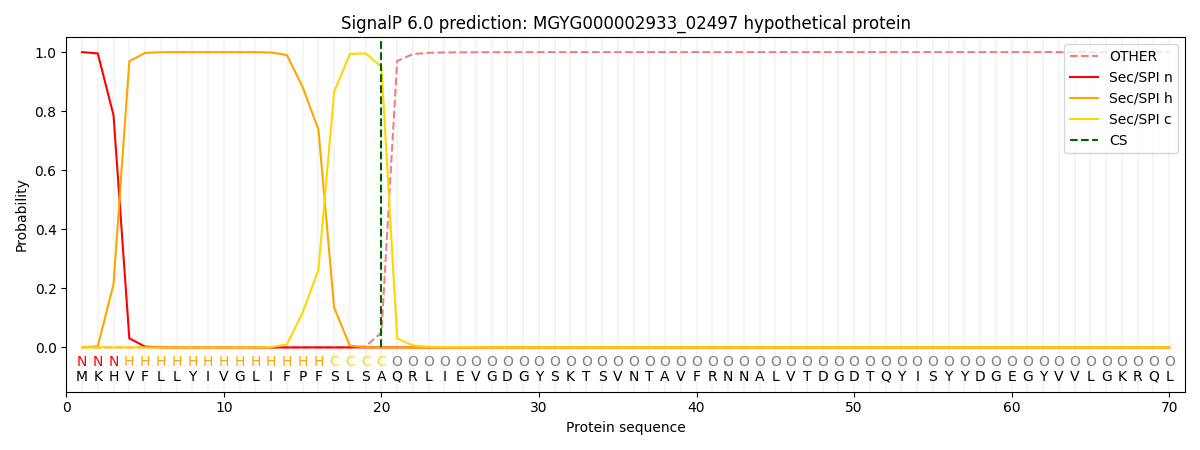
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000467 | 0.998507 | 0.000453 | 0.000181 | 0.000178 | 0.000175 |
