You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002945_02543
You are here: Home > Sequence: MGYG000002945_02543
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
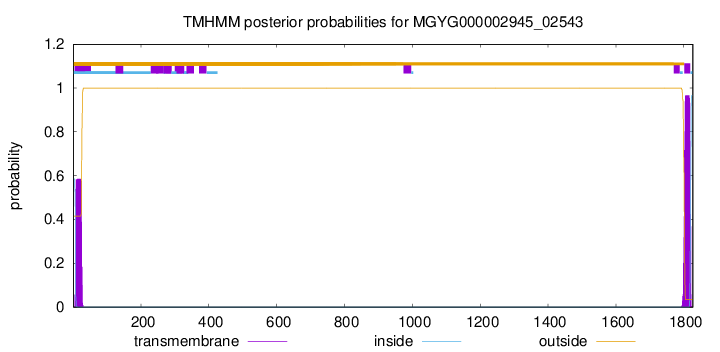
TMHMM annotations
Basic Information help
| Species | Bariatricus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bariatricus; Bariatricus massiliensis | |||||||||||
| CAZyme ID | MGYG000002945_02543 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73309; End: 78792 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 337 | 1051 | 6e-64 | 0.7300531914893617 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.32e-19 | 336 | 774 | 1 | 410 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02368 | Big_2 | 8.41e-19 | 1671 | 1747 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG5492 | YjdB | 1.73e-18 | 1668 | 1750 | 179 | 262 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| PRK10150 | PRK10150 | 2.89e-18 | 341 | 662 | 5 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.13e-16 | 350 | 770 | 44 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANX00219.1 | 2.41e-185 | 346 | 1253 | 4 | 902 |
| AGI38265.1 | 2.41e-185 | 346 | 1253 | 4 | 902 |
| ANW97658.1 | 2.41e-185 | 346 | 1253 | 4 | 902 |
| AGC67186.1 | 2.41e-185 | 346 | 1253 | 4 | 902 |
| BBE17052.1 | 2.45e-184 | 347 | 1270 | 29 | 953 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 2.59e-15 | 407 | 804 | 90 | 465 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6U7I_A | 3.61e-10 | 348 | 791 | 13 | 439 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 2V72_A | 6.86e-08 | 45 | 182 | 12 | 142 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 2VZO_A | 1.10e-07 | 431 | 773 | 146 | 472 | Crystalstructure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZS_A | 2.47e-07 | 431 | 773 | 146 | 472 | ChitosanProduct complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0KQH4 | 6.03e-11 | 407 | 770 | 126 | 462 | Beta-galactosidase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=lacZ PE=3 SV=1 |
| P85991 | 3.56e-09 | 1632 | 1750 | 145 | 258 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
| P35838 | 3.57e-09 | 1671 | 1757 | 237 | 324 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
| A0A3R0A696 | 1.37e-08 | 1668 | 1749 | 270 | 351 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| Q48846 | 1.83e-08 | 401 | 774 | 127 | 469 | Beta-galactosidase large subunit OS=Latilactobacillus sakei OX=1599 GN=lacL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000265 | 0.999038 | 0.000196 | 0.000177 | 0.000151 | 0.000133 |