You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002960_00935
You are here: Home > Sequence: MGYG000002960_00935
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900555035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900555035 | |||||||||||
| CAZyme ID | MGYG000002960_00935 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24177; End: 25676 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 50 | 496 | 8.6e-155 | 0.9928057553956835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02055 | Glyco_hydro_30 | 4.58e-31 | 64 | 432 | 4 | 348 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| COG5520 | XynC | 2.86e-20 | 1 | 499 | 1 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT62831.1 | 7.09e-226 | 20 | 496 | 28 | 503 |
| QUT58833.1 | 7.09e-226 | 20 | 496 | 28 | 503 |
| QUT68391.1 | 7.09e-226 | 20 | 496 | 28 | 503 |
| QQA31246.1 | 7.09e-226 | 20 | 496 | 28 | 503 |
| ABR39831.1 | 7.09e-226 | 20 | 496 | 28 | 503 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WNW_A | 1.22e-25 | 64 | 499 | 48 | 443 | Thecrystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2WNW_B The crystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5NGK_A | 2.38e-16 | 76 | 498 | 92 | 490 | Theendo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O16580 | 3.97e-24 | 46 | 436 | 83 | 449 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
| Q9UB00 | 5.95e-21 | 41 | 395 | 81 | 409 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
| G5ECR8 | 1.93e-20 | 48 | 499 | 91 | 519 | Putative glucosylceramidase 3 OS=Caenorhabditis elegans OX=6239 GN=gba-3 PE=3 SV=1 |
| P17439 | 2.55e-18 | 48 | 499 | 84 | 513 | Lysosomal acid glucosylceramidase OS=Mus musculus OX=10090 GN=Gba PE=1 SV=1 |
| Q4WBR2 | 1.14e-11 | 48 | 333 | 60 | 311 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
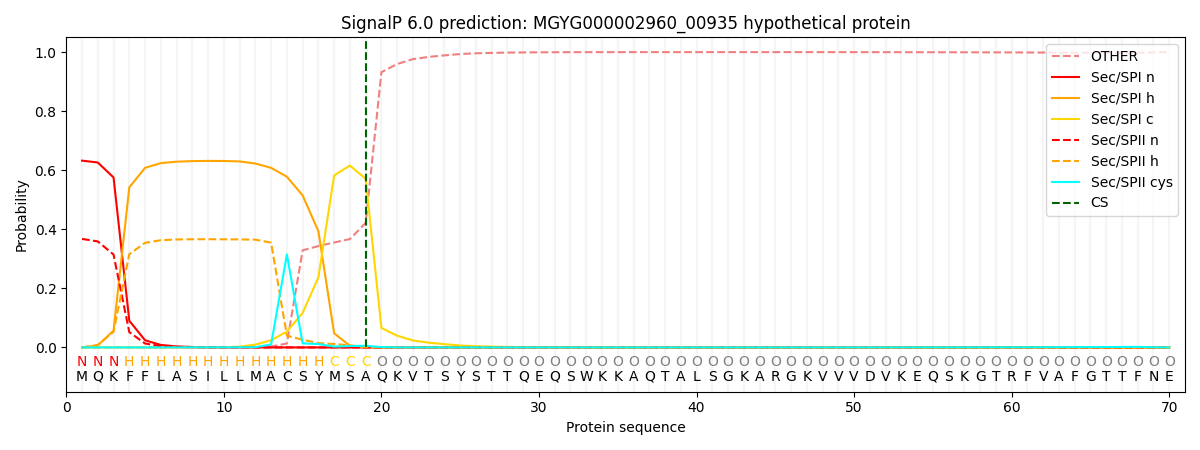
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000721 | 0.621674 | 0.376860 | 0.000242 | 0.000252 | 0.000240 |
