You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002960_01169
You are here: Home > Sequence: MGYG000002960_01169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900555035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900555035 | |||||||||||
| CAZyme ID | MGYG000002960_01169 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 743; End: 3988 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 642 | 869 | 2.1e-40 | 0.7673611111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.28e-15 | 638 | 856 | 87 | 332 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PRK10531 | PRK10531 | 6.18e-12 | 711 | 862 | 193 | 354 | putative acyl-CoA thioester hydrolase. |
| PLN02488 | PLN02488 | 1.69e-09 | 663 | 866 | 226 | 428 | probable pectinesterase/pectinesterase inhibitor |
| PLN02217 | PLN02217 | 3.80e-09 | 684 | 868 | 298 | 483 | probable pectinesterase/pectinesterase inhibitor |
| PLN02708 | PLN02708 | 1.02e-08 | 636 | 867 | 244 | 483 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH16322.1 | 1.74e-225 | 159 | 1081 | 562 | 1471 |
| AGB28243.1 | 4.04e-198 | 157 | 1081 | 558 | 1371 |
| QUT74167.1 | 2.22e-109 | 22 | 1081 | 262 | 1435 |
| QCD38476.1 | 3.41e-79 | 524 | 1081 | 942 | 1476 |
| QCP72166.1 | 3.41e-79 | 524 | 1081 | 942 | 1476 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P55743 | 4.68e-10 | 715 | 862 | 175 | 334 | Pectinesterase B OS=Pectobacterium parmentieri OX=1905730 GN=pemB PE=3 SV=2 |
| Q3EAY9 | 2.62e-09 | 652 | 868 | 202 | 418 | Probable pectinesterase 30 OS=Arabidopsis thaliana OX=3702 GN=PME30 PE=2 SV=1 |
| P83947 | 2.88e-09 | 687 | 869 | 282 | 465 | Pectinesterase/pectinesterase inhibitor OS=Ficus pumila var. awkeotsang OX=204231 PE=1 SV=1 |
| Q47474 | 2.05e-08 | 711 | 856 | 200 | 359 | Pectinesterase B OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemB PE=1 SV=2 |
| Q84JX1 | 5.77e-08 | 652 | 855 | 229 | 432 | Probable pectinesterase/pectinesterase inhibitor 19 OS=Arabidopsis thaliana OX=3702 GN=PME19 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
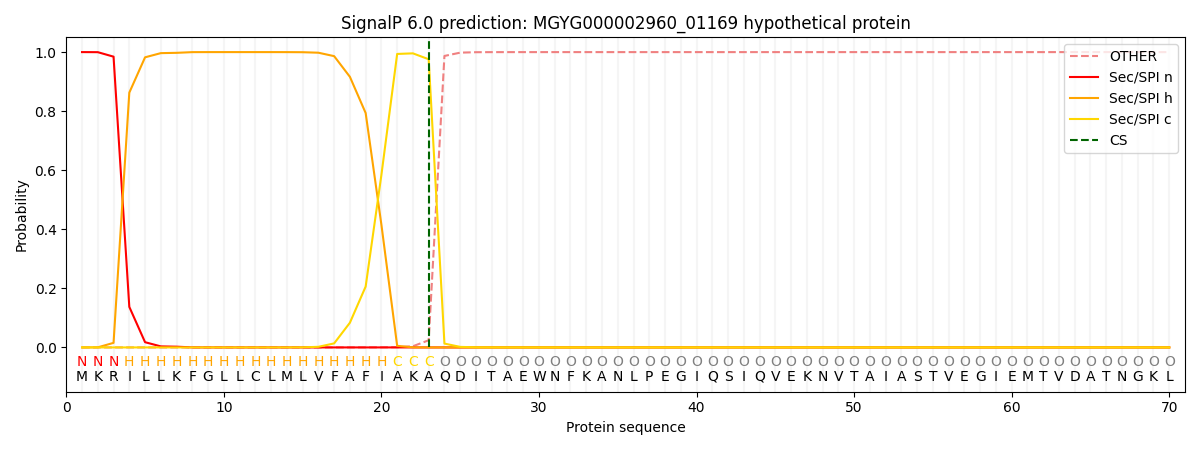
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000250 | 0.999105 | 0.000188 | 0.000158 | 0.000144 | 0.000144 |
