You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002963_02416
You are here: Home > Sequence: MGYG000002963_02416
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
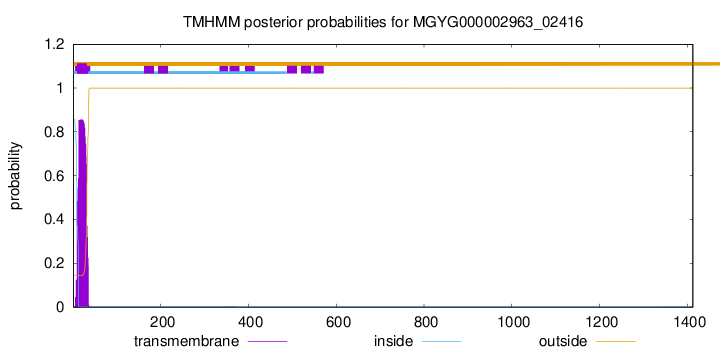
TMHMM annotations
Basic Information help
| Species | Anaerosporobacter mobilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerosporobacter; Anaerosporobacter mobilis | |||||||||||
| CAZyme ID | MGYG000002963_02416 | |||||||||||
| CAZy Family | CBM27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55723; End: 59964 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 427 | 783 | 2.5e-50 | 0.9174917491749175 |
| CBM23 | 947 | 1104 | 3.8e-30 | 0.9135802469135802 |
| CBM27 | 211 | 373 | 8.4e-17 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 2.93e-31 | 427 | 707 | 1 | 241 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.54e-26 | 590 | 782 | 157 | 343 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| sd00036 | LRR_3 | 2.03e-25 | 1279 | 1391 | 14 | 116 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 3.31e-24 | 1298 | 1391 | 1 | 93 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 1.20e-22 | 1300 | 1391 | 1 | 89 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQQ93186.1 | 2.39e-233 | 165 | 1134 | 181 | 1118 |
| ASU28432.1 | 9.58e-233 | 165 | 1134 | 228 | 1165 |
| ANU75629.1 | 9.58e-233 | 165 | 1134 | 228 | 1165 |
| QJU14275.1 | 5.15e-232 | 165 | 1134 | 181 | 1118 |
| AEY67866.1 | 8.03e-228 | 75 | 1133 | 255 | 1258 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 4.90e-57 | 425 | 822 | 50 | 416 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1J9Y_A | 5.91e-45 | 424 | 820 | 9 | 369 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1R7O_A | 7.47e-45 | 424 | 820 | 19 | 379 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 1GW1_A | 2.96e-44 | 424 | 820 | 5 | 362 | Substratedistortion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1GVY_A | 3.49e-44 | 424 | 820 | 9 | 369 | Substratedistorsion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1A278 | 2.23e-48 | 424 | 1069 | 40 | 641 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49424 | 7.71e-44 | 424 | 820 | 47 | 407 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| P16699 | 1.07e-14 | 426 | 703 | 34 | 279 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| P49425 | 1.52e-13 | 601 | 734 | 289 | 408 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
| P55278 | 1.66e-12 | 574 | 719 | 140 | 291 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000375 | 0.998767 | 0.000229 | 0.000239 | 0.000191 | 0.000171 |