You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002963_03151
You are here: Home > Sequence: MGYG000002963_03151
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerosporobacter mobilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerosporobacter; Anaerosporobacter mobilis | |||||||||||
| CAZyme ID | MGYG000002963_03151 | |||||||||||
| CAZy Family | GH74 | |||||||||||
| CAZyme Description | Xyloglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73655; End: 76792 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM3 | 903 | 983 | 1e-24 | 0.9772727272727273 |
| GH74 | 99 | 208 | 3e-23 | 0.4592274678111588 |
| GH74 | 610 | 735 | 1.3e-18 | 0.48068669527896996 |
| CBM46 | 817 | 887 | 9.7e-18 | 0.8620689655172413 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03442 | CBM_X2 | 1.82e-32 | 805 | 887 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| pfam00942 | CBM_3 | 2.60e-22 | 902 | 982 | 1 | 81 | Cellulose binding domain. |
| smart01067 | CBM_3 | 1.11e-20 | 902 | 984 | 1 | 83 | Cellulose binding domain. |
| pfam15902 | Sortilin-Vps10 | 1.45e-05 | 105 | 210 | 21 | 118 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| pfam15902 | Sortilin-Vps10 | 6.09e-05 | 592 | 707 | 2 | 112 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQZ59424.1 | 0.0 | 44 | 1045 | 36 | 1032 |
| QMV44280.1 | 0.0 | 20 | 1045 | 19 | 1122 |
| CQR55184.1 | 0.0 | 44 | 1045 | 36 | 1127 |
| ATB38151.1 | 0.0 | 44 | 1045 | 40 | 1029 |
| QRK14121.1 | 0.0 | 47 | 1045 | 43 | 1026 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MGJ_A | 0.0 | 44 | 794 | 3 | 747 | Crystalstructure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_B Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_C Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_D Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_E Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_F Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_G Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGJ_H Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme [Paenibacillus odorifer],6MGK_A Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_B Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_C Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer],6MGK_D Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan [Paenibacillus odorifer] |
| 6P2N_A | 0.0 | 44 | 794 | 3 | 748 | Crystalstructure of Paenibacillus graminis GH74 (PgGH74) [Paenibacillus graminis] |
| 6MGL_A | 0.0 | 44 | 794 | 3 | 747 | Crystalstructure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan [Paenibacillus odorifer] |
| 4LGN_A | 1.47e-284 | 44 | 792 | 4 | 734 | Thestructure of Acidothermus cellulolyticus family 74 glycoside hydrolase [Acidothermus cellulolyticus 11B] |
| 2CN3_A | 1.63e-280 | 44 | 798 | 8 | 736 | ChainA, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus],2CN3_B Chain B, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3MUH7 | 0.0 | 44 | 1045 | 35 | 1027 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
| A3DFA0 | 4.12e-282 | 8 | 798 | 3 | 763 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xghA PE=3 SV=1 |
| Q70DK5 | 1.65e-281 | 8 | 798 | 3 | 763 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus OX=1515 GN=xghA PE=1 SV=1 |
| Q7Z9M8 | 4.80e-186 | 47 | 790 | 21 | 742 | Xyloglucanase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cel74a PE=1 SV=1 |
| Q5BD38 | 5.54e-132 | 44 | 792 | 32 | 807 | Oligoxyloglucan-reducing end-specific xyloglucanase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgcA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
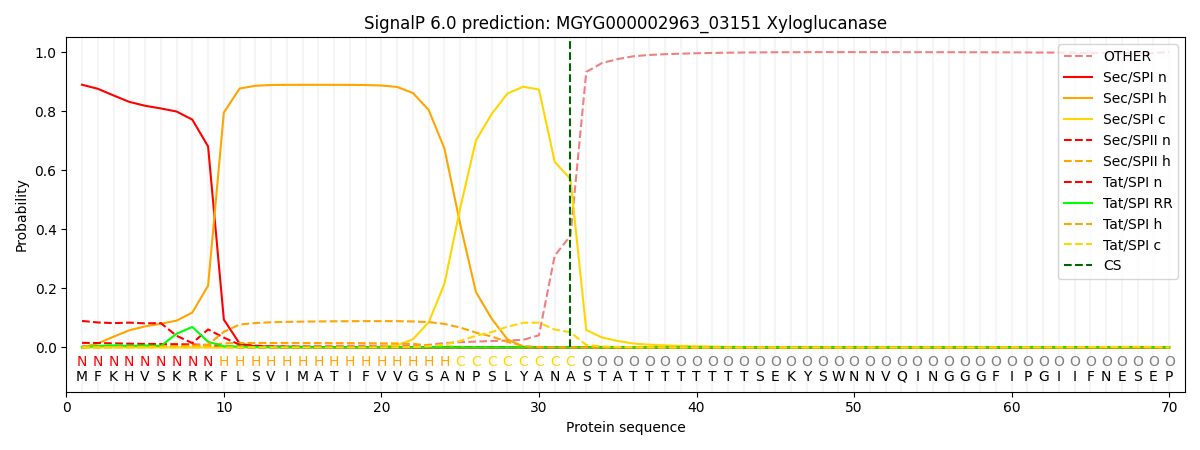
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007114 | 0.881929 | 0.017349 | 0.091657 | 0.001592 | 0.000322 |
