You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002977_00246
You are here: Home > Sequence: MGYG000002977_00246
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
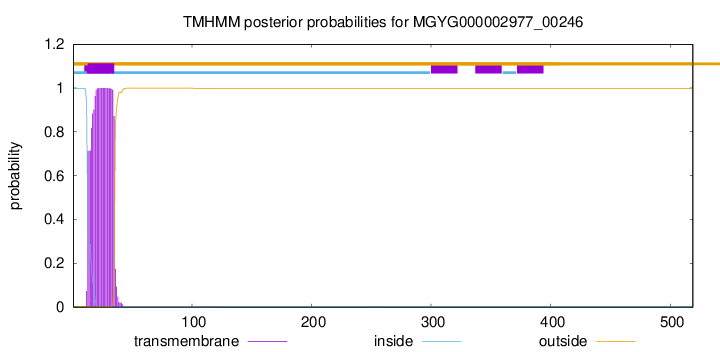
TMHMM annotations
Basic Information help
| Species | Bifidobacterium vaginale_F | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium vaginale_F | |||||||||||
| CAZyme ID | MGYG000002977_00246 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1997; End: 3556 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3583 | YabE | 2.82e-35 | 17 | 287 | 16 | 288 | Uncharacterized conserved protein YabE, contains G5 and tandem DUF348 domains [Function unknown]. |
| pfam07501 | G5 | 1.06e-17 | 211 | 283 | 3 | 75 | G5 domain. This domain is found in a wide range of extracellular proteins. It is found tandemly repeated in up to 8 copies. It is found in the N-terminus of peptidases belonging to the M26 family which cleave human IgA. The domain is also found in proteins involved in metabolism of bacterial cell walls suggesting this domain may have an adhesive function. |
| pfam03990 | DUF348 | 9.78e-05 | 41 | 79 | 1 | 39 | Domain of unknown function (DUF348). This domain normally occurs as tandem repeats; however it is found as a single copy in the S. cerevisiae DNA-binding nuclear protein YCR593. This protein is involved in sporulation part of the SET3C complex, which is required to repress early/middle sporulation genes during meiosis. The bacterial proteins are likely to be involved in a cell wall function as they are found in conjunction with the pfam07501 domain, which is involved in various cell surface processes. |
| cd13925 | RPF | 7.98e-04 | 436 | 492 | 2 | 53 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
| pfam03990 | DUF348 | 0.004 | 156 | 193 | 1 | 38 | Domain of unknown function (DUF348). This domain normally occurs as tandem repeats; however it is found as a single copy in the S. cerevisiae DNA-binding nuclear protein YCR593. This protein is involved in sporulation part of the SET3C complex, which is required to repress early/middle sporulation genes during meiosis. The bacterial proteins are likely to be involved in a cell wall function as they are found in conjunction with the pfam07501 domain, which is involved in various cell surface processes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYZ21238.1 | 1.48e-180 | 1 | 519 | 1 | 504 |
| SDR85797.1 | 1.58e-178 | 1 | 519 | 1 | 508 |
| ADP38195.1 | 1.58e-178 | 1 | 519 | 1 | 508 |
| VEH17271.1 | 1.58e-178 | 1 | 519 | 1 | 508 |
| ADB14393.1 | 9.88e-178 | 1 | 519 | 1 | 460 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6M6N7 | 4.45e-12 | 31 | 267 | 35 | 273 | Resuscitation-promoting factor Rpf2 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=rpf2 PE=1 SV=1 |
| P37546 | 5.69e-06 | 34 | 163 | 90 | 219 | Putative cell wall shaping protein YabE OS=Bacillus subtilis (strain 168) OX=224308 GN=yabE PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
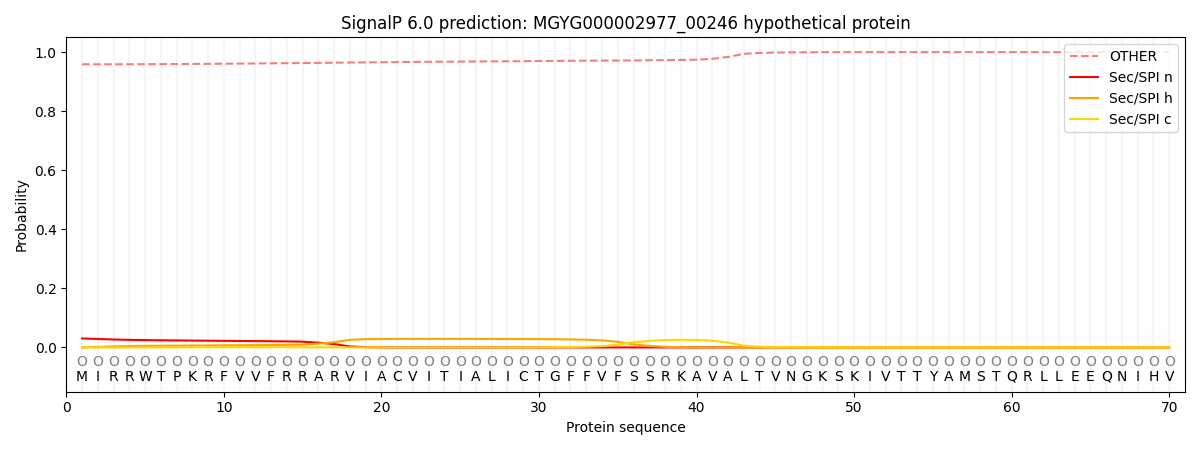
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.960295 | 0.028134 | 0.008651 | 0.000117 | 0.000081 | 0.002737 |