You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002977_00621
Basic Information
help
| Species |
Bifidobacterium vaginale_F
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium vaginale_F
|
| CAZyme ID |
MGYG000002977_00621
|
| CAZy Family |
GT2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002977 |
1471933 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 3503;
End: 6865
Strand: -
|
No EC number prediction in MGYG000002977_00621.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1216
|
GT2 |
0.002 |
142 |
364 |
70 |
281 |
Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000065
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
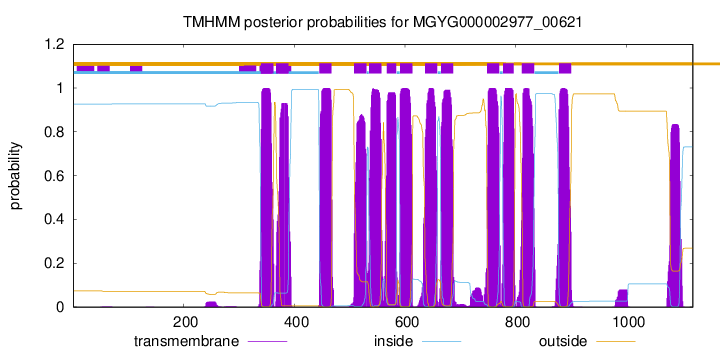
| start |
end |
| 340 |
362 |
| 367 |
389 |
| 445 |
467 |
| 508 |
530 |
| 535 |
557 |
| 567 |
584 |
| 591 |
613 |
| 636 |
658 |
| 665 |
687 |
| 748 |
770 |
| 777 |
796 |
| 811 |
833 |
| 878 |
900 |


