You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002993_02126
You are here: Home > Sequence: MGYG000002993_02126
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
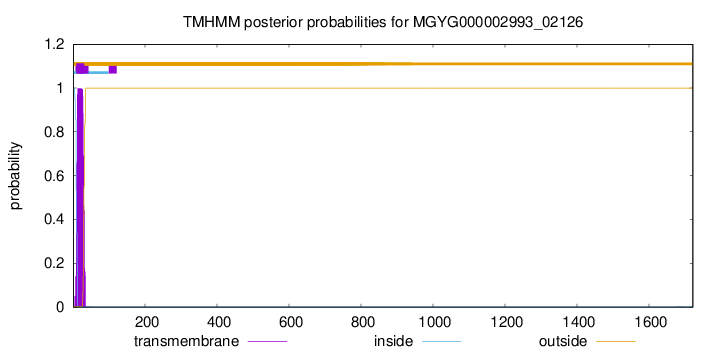
TMHMM annotations
Basic Information help
| Species | CAG-170 sp000432135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-170; CAG-170 sp000432135 | |||||||||||
| CAZyme ID | MGYG000002993_02126 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3034; End: 8202 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1079 | 1208 | 4.3e-19 | 0.9112903225806451 |
| CBM32 | 1250 | 1381 | 5.2e-17 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.07e-13 | 409 | 540 | 2 | 116 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 6.10e-13 | 1081 | 1211 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 8.19e-12 | 295 | 373 | 39 | 117 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02368 | Big_2 | 2.77e-10 | 567 | 631 | 2 | 66 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam00754 | F5_F8_type_C | 1.12e-09 | 1260 | 1364 | 11 | 112 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDS28274.1 | 1.77e-136 | 365 | 1062 | 29 | 694 |
| QOR71214.1 | 1.60e-120 | 224 | 1055 | 44 | 838 |
| QBR87893.1 | 9.96e-119 | 220 | 1055 | 38 | 829 |
| SDS02659.1 | 1.30e-100 | 213 | 1218 | 18 | 1024 |
| SDS02613.1 | 5.11e-92 | 228 | 1218 | 36 | 1021 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 1.25e-11 | 1549 | 1720 | 1275 | 1461 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38536 | 4.29e-08 | 1557 | 1719 | 1682 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P35838 | 2.24e-07 | 571 | 690 | 242 | 355 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
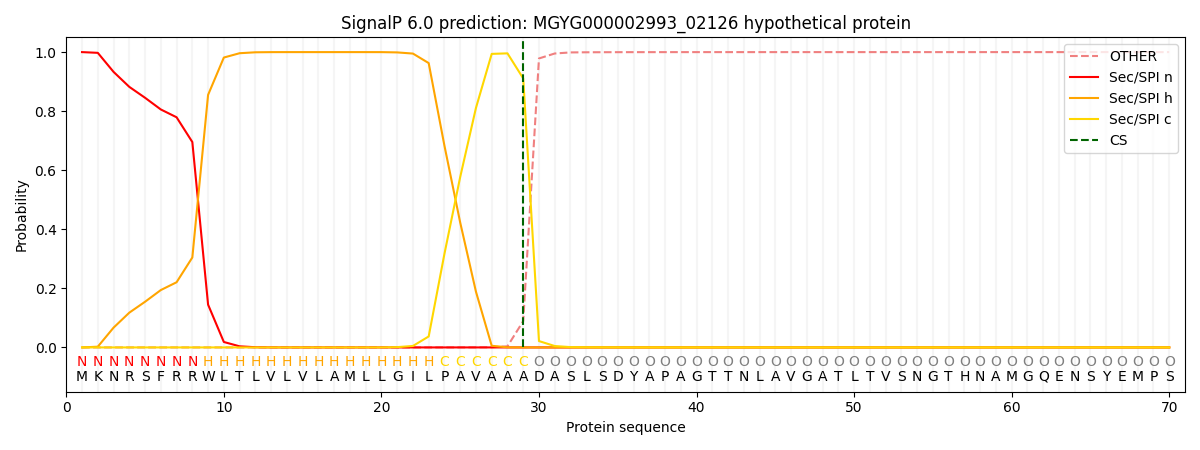
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000290 | 0.998936 | 0.000221 | 0.000199 | 0.000178 | 0.000157 |